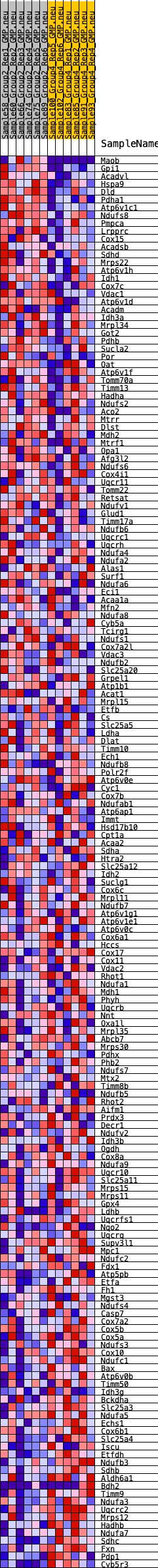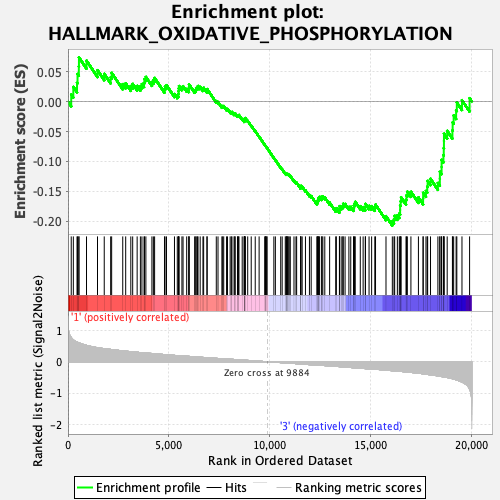
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | GMP.GMP.neu_Pheno.cls #Group2_versus_Group4.GMP.neu_Pheno.cls #Group2_versus_Group4_repos |
| Phenotype | GMP.neu_Pheno.cls#Group2_versus_Group4_repos |
| Upregulated in class | 3 |
| GeneSet | HALLMARK_OXIDATIVE_PHOSPHORYLATION |
| Enrichment Score (ES) | -0.20696786 |
| Normalized Enrichment Score (NES) | -0.7849053 |
| Nominal p-value | 0.65731466 |
| FDR q-value | 0.8297606 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Maob | 160 | 0.775 | 0.0118 | No |
| 2 | Gpi1 | 264 | 0.697 | 0.0244 | No |
| 3 | Acadvl | 448 | 0.629 | 0.0313 | No |
| 4 | Hspa9 | 473 | 0.622 | 0.0460 | No |
| 5 | Dld | 537 | 0.608 | 0.0583 | No |
| 6 | Pdha1 | 543 | 0.606 | 0.0736 | No |
| 7 | Atp6v1c1 | 918 | 0.524 | 0.0681 | No |
| 8 | Ndufs8 | 1463 | 0.451 | 0.0523 | No |
| 9 | Pmpca | 1797 | 0.418 | 0.0462 | No |
| 10 | Lrpprc | 2116 | 0.395 | 0.0403 | No |
| 11 | Cox15 | 2165 | 0.390 | 0.0478 | No |
| 12 | Acadsb | 2715 | 0.349 | 0.0291 | No |
| 13 | Sdhd | 2861 | 0.340 | 0.0305 | No |
| 14 | Mrps22 | 3115 | 0.322 | 0.0259 | No |
| 15 | Atp6v1h | 3208 | 0.316 | 0.0294 | No |
| 16 | Idh1 | 3429 | 0.304 | 0.0261 | No |
| 17 | Cox7c | 3590 | 0.294 | 0.0255 | No |
| 18 | Vdac1 | 3666 | 0.291 | 0.0292 | No |
| 19 | Atp6v1d | 3771 | 0.284 | 0.0312 | No |
| 20 | Acadm | 3794 | 0.282 | 0.0373 | No |
| 21 | Idh3a | 3862 | 0.279 | 0.0411 | No |
| 22 | Mrpl34 | 4162 | 0.265 | 0.0328 | No |
| 23 | Got2 | 4234 | 0.259 | 0.0359 | No |
| 24 | Pdhb | 4295 | 0.255 | 0.0394 | No |
| 25 | Sucla2 | 4785 | 0.229 | 0.0206 | No |
| 26 | Por | 4812 | 0.228 | 0.0251 | No |
| 27 | Oat | 4884 | 0.223 | 0.0272 | No |
| 28 | Atp6v1f | 5280 | 0.204 | 0.0125 | No |
| 29 | Tomm70a | 5420 | 0.196 | 0.0105 | No |
| 30 | Timm13 | 5478 | 0.192 | 0.0126 | No |
| 31 | Hadha | 5484 | 0.192 | 0.0172 | No |
| 32 | Ndufs2 | 5490 | 0.191 | 0.0219 | No |
| 33 | Aco2 | 5508 | 0.190 | 0.0259 | No |
| 34 | Mtrr | 5650 | 0.189 | 0.0236 | No |
| 35 | Dlst | 5705 | 0.185 | 0.0256 | No |
| 36 | Mdh2 | 5886 | 0.176 | 0.0210 | No |
| 37 | Mtrf1 | 5995 | 0.172 | 0.0200 | No |
| 38 | Opa1 | 6000 | 0.172 | 0.0242 | No |
| 39 | Afg3l2 | 6002 | 0.172 | 0.0285 | No |
| 40 | Ndufs6 | 6280 | 0.158 | 0.0186 | No |
| 41 | Cox4i1 | 6335 | 0.155 | 0.0199 | No |
| 42 | Uqcr11 | 6358 | 0.154 | 0.0227 | No |
| 43 | Tomm22 | 6420 | 0.151 | 0.0235 | No |
| 44 | Retsat | 6448 | 0.150 | 0.0260 | No |
| 45 | Ndufv1 | 6555 | 0.147 | 0.0244 | No |
| 46 | Glud1 | 6693 | 0.139 | 0.0210 | No |
| 47 | Timm17a | 6727 | 0.137 | 0.0229 | No |
| 48 | Ndufb6 | 6891 | 0.128 | 0.0180 | No |
| 49 | Uqcrc1 | 6900 | 0.128 | 0.0208 | No |
| 50 | Uqcrh | 7354 | 0.109 | 0.0008 | No |
| 51 | Ndufa4 | 7439 | 0.105 | -0.0008 | No |
| 52 | Ndufa2 | 7632 | 0.096 | -0.0080 | No |
| 53 | Alas1 | 7670 | 0.095 | -0.0074 | No |
| 54 | Surf1 | 7722 | 0.092 | -0.0076 | No |
| 55 | Ndufa6 | 7863 | 0.090 | -0.0124 | No |
| 56 | Eci1 | 7909 | 0.088 | -0.0124 | No |
| 57 | Acaa1a | 8041 | 0.081 | -0.0169 | No |
| 58 | Mfn2 | 8100 | 0.079 | -0.0178 | No |
| 59 | Ndufa8 | 8131 | 0.078 | -0.0173 | No |
| 60 | Cyb5a | 8235 | 0.073 | -0.0207 | No |
| 61 | Tcirg1 | 8271 | 0.071 | -0.0206 | No |
| 62 | Ndufs1 | 8290 | 0.070 | -0.0197 | No |
| 63 | Cox7a2l | 8405 | 0.065 | -0.0238 | No |
| 64 | Vdac3 | 8422 | 0.064 | -0.0230 | No |
| 65 | Ndufb2 | 8460 | 0.063 | -0.0232 | No |
| 66 | Slc25a20 | 8476 | 0.062 | -0.0224 | No |
| 67 | Grpel1 | 8639 | 0.056 | -0.0291 | No |
| 68 | Atp1b1 | 8740 | 0.051 | -0.0329 | No |
| 69 | Acat1 | 8742 | 0.051 | -0.0316 | No |
| 70 | Mrpl15 | 8749 | 0.051 | -0.0306 | No |
| 71 | Etfb | 8750 | 0.051 | -0.0293 | No |
| 72 | Cs | 8766 | 0.050 | -0.0288 | No |
| 73 | Slc25a5 | 8776 | 0.050 | -0.0279 | No |
| 74 | Ldha | 8794 | 0.049 | -0.0275 | No |
| 75 | Dlat | 8912 | 0.044 | -0.0323 | No |
| 76 | Timm10 | 9086 | 0.036 | -0.0401 | No |
| 77 | Ech1 | 9285 | 0.028 | -0.0494 | No |
| 78 | Ndufb8 | 9478 | 0.019 | -0.0586 | No |
| 79 | Polr2f | 9755 | 0.006 | -0.0723 | No |
| 80 | Atp6v0e | 9790 | 0.005 | -0.0739 | No |
| 81 | Cyc1 | 9798 | 0.005 | -0.0741 | No |
| 82 | Cox7b | 9813 | 0.004 | -0.0747 | No |
| 83 | Ndufab1 | 9851 | 0.002 | -0.0766 | No |
| 84 | Atp6ap1 | 9873 | 0.001 | -0.0776 | No |
| 85 | Immt | 10209 | -0.011 | -0.0942 | No |
| 86 | Hsd17b10 | 10284 | -0.014 | -0.0976 | No |
| 87 | Cpt1a | 10549 | -0.025 | -0.1102 | No |
| 88 | Acaa2 | 10621 | -0.028 | -0.1131 | No |
| 89 | Sdha | 10775 | -0.033 | -0.1199 | No |
| 90 | Htra2 | 10824 | -0.035 | -0.1215 | No |
| 91 | Slc25a12 | 10825 | -0.035 | -0.1205 | No |
| 92 | Idh2 | 10842 | -0.036 | -0.1204 | No |
| 93 | Suclg1 | 10877 | -0.038 | -0.1212 | No |
| 94 | Cox6c | 10896 | -0.039 | -0.1211 | No |
| 95 | Mrpl11 | 10921 | -0.040 | -0.1213 | No |
| 96 | Ndufb7 | 10970 | -0.043 | -0.1226 | No |
| 97 | Atp6v1g1 | 11024 | -0.044 | -0.1241 | No |
| 98 | Atp6v1e1 | 11207 | -0.053 | -0.1319 | No |
| 99 | Atp6v0c | 11306 | -0.057 | -0.1354 | No |
| 100 | Cox6a1 | 11339 | -0.059 | -0.1355 | No |
| 101 | Hccs | 11535 | -0.068 | -0.1436 | No |
| 102 | Cox17 | 11545 | -0.069 | -0.1423 | No |
| 103 | Cox11 | 11547 | -0.069 | -0.1406 | No |
| 104 | Vdac2 | 11612 | -0.072 | -0.1420 | No |
| 105 | Rhot1 | 11776 | -0.079 | -0.1482 | No |
| 106 | Ndufa1 | 11973 | -0.089 | -0.1558 | No |
| 107 | Mdh1 | 12060 | -0.093 | -0.1577 | No |
| 108 | Phyh | 12342 | -0.105 | -0.1692 | No |
| 109 | Uqcrb | 12368 | -0.107 | -0.1677 | No |
| 110 | Nnt | 12379 | -0.107 | -0.1655 | No |
| 111 | Oxa1l | 12407 | -0.108 | -0.1640 | No |
| 112 | Mrpl35 | 12417 | -0.109 | -0.1617 | No |
| 113 | Abcb7 | 12443 | -0.110 | -0.1602 | No |
| 114 | Mrps30 | 12478 | -0.111 | -0.1591 | No |
| 115 | Pdhx | 12584 | -0.114 | -0.1614 | No |
| 116 | Phb2 | 12587 | -0.114 | -0.1586 | No |
| 117 | Ndufs7 | 12638 | -0.116 | -0.1582 | No |
| 118 | Mtx2 | 12735 | -0.120 | -0.1599 | No |
| 119 | Timm8b | 12975 | -0.131 | -0.1686 | No |
| 120 | Ndufb5 | 13277 | -0.144 | -0.1801 | No |
| 121 | Rhot2 | 13309 | -0.146 | -0.1779 | No |
| 122 | Aifm1 | 13460 | -0.153 | -0.1816 | No |
| 123 | Prdx3 | 13470 | -0.154 | -0.1781 | No |
| 124 | Decr1 | 13476 | -0.154 | -0.1744 | No |
| 125 | Ndufv2 | 13581 | -0.160 | -0.1756 | No |
| 126 | Idh3b | 13634 | -0.163 | -0.1740 | No |
| 127 | Ogdh | 13647 | -0.164 | -0.1705 | No |
| 128 | Cox8a | 13741 | -0.168 | -0.1708 | No |
| 129 | Ndufa9 | 13930 | -0.178 | -0.1758 | No |
| 130 | Uqcr10 | 14022 | -0.182 | -0.1757 | No |
| 131 | Slc25a11 | 14159 | -0.189 | -0.1777 | No |
| 132 | Mrps15 | 14176 | -0.189 | -0.1737 | No |
| 133 | Mrps11 | 14207 | -0.191 | -0.1703 | No |
| 134 | Gpx4 | 14245 | -0.192 | -0.1672 | No |
| 135 | Ldhb | 14495 | -0.204 | -0.1746 | No |
| 136 | Uqcrfs1 | 14636 | -0.212 | -0.1762 | No |
| 137 | Nqo2 | 14745 | -0.217 | -0.1761 | No |
| 138 | Uqcrq | 14750 | -0.217 | -0.1708 | No |
| 139 | Supv3l1 | 14930 | -0.226 | -0.1740 | No |
| 140 | Mpc1 | 15058 | -0.232 | -0.1744 | No |
| 141 | Ndufc2 | 15216 | -0.237 | -0.1763 | No |
| 142 | Fdx1 | 15251 | -0.239 | -0.1719 | No |
| 143 | Atp5pb | 15770 | -0.266 | -0.1912 | No |
| 144 | Etfa | 16084 | -0.283 | -0.1997 | Yes |
| 145 | Fh1 | 16166 | -0.288 | -0.1964 | Yes |
| 146 | Mgst3 | 16200 | -0.290 | -0.1907 | Yes |
| 147 | Ndufs4 | 16334 | -0.298 | -0.1898 | Yes |
| 148 | Casp7 | 16429 | -0.304 | -0.1867 | Yes |
| 149 | Cox7a2 | 16471 | -0.306 | -0.1810 | Yes |
| 150 | Cox5b | 16472 | -0.306 | -0.1732 | Yes |
| 151 | Cox5a | 16497 | -0.307 | -0.1665 | Yes |
| 152 | Ndufs3 | 16519 | -0.308 | -0.1597 | Yes |
| 153 | Cox10 | 16764 | -0.322 | -0.1637 | Yes |
| 154 | Ndufc1 | 16786 | -0.324 | -0.1565 | Yes |
| 155 | Bax | 16829 | -0.326 | -0.1503 | Yes |
| 156 | Atp6v0b | 17006 | -0.338 | -0.1505 | Yes |
| 157 | Timm50 | 17378 | -0.364 | -0.1599 | Yes |
| 158 | Idh3g | 17610 | -0.383 | -0.1618 | Yes |
| 159 | Bckdha | 17613 | -0.383 | -0.1521 | Yes |
| 160 | Slc25a3 | 17753 | -0.396 | -0.1490 | Yes |
| 161 | Ndufa5 | 17816 | -0.403 | -0.1418 | Yes |
| 162 | Echs1 | 17836 | -0.405 | -0.1324 | Yes |
| 163 | Cox6b1 | 17978 | -0.414 | -0.1289 | Yes |
| 164 | Slc25a4 | 18344 | -0.450 | -0.1358 | Yes |
| 165 | Iscu | 18441 | -0.460 | -0.1289 | Yes |
| 166 | Etfdh | 18443 | -0.461 | -0.1171 | Yes |
| 167 | Ndufb3 | 18527 | -0.471 | -0.1093 | Yes |
| 168 | Sdhb | 18529 | -0.471 | -0.0972 | Yes |
| 169 | Aldh6a1 | 18619 | -0.481 | -0.0894 | Yes |
| 170 | Bdh2 | 18635 | -0.481 | -0.0779 | Yes |
| 171 | Timm9 | 18644 | -0.481 | -0.0660 | Yes |
| 172 | Ndufa3 | 18646 | -0.482 | -0.0537 | Yes |
| 173 | Uqcrc2 | 18805 | -0.499 | -0.0489 | Yes |
| 174 | Mrps12 | 19064 | -0.539 | -0.0481 | Yes |
| 175 | Hadhb | 19076 | -0.542 | -0.0348 | Yes |
| 176 | Ndufa7 | 19126 | -0.551 | -0.0232 | Yes |
| 177 | Sdhc | 19248 | -0.574 | -0.0146 | Yes |
| 178 | Fxn | 19283 | -0.580 | -0.0015 | Yes |
| 179 | Pdp1 | 19538 | -0.646 | 0.0023 | Yes |
| 180 | Cyb5r3 | 19916 | -0.871 | 0.0055 | Yes |