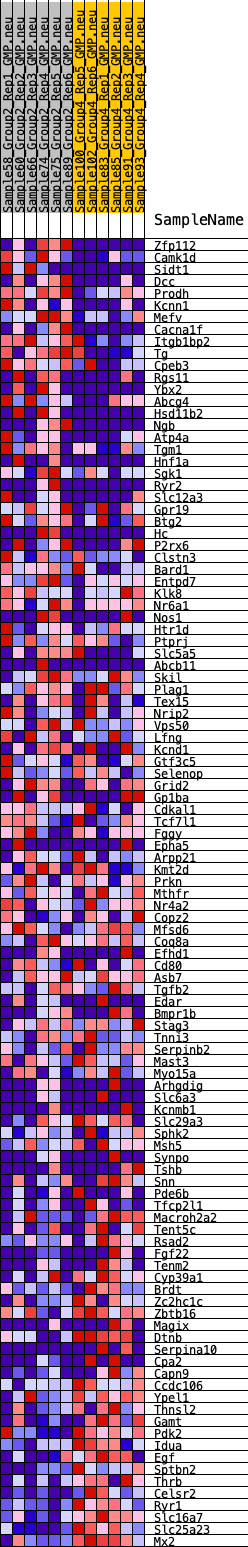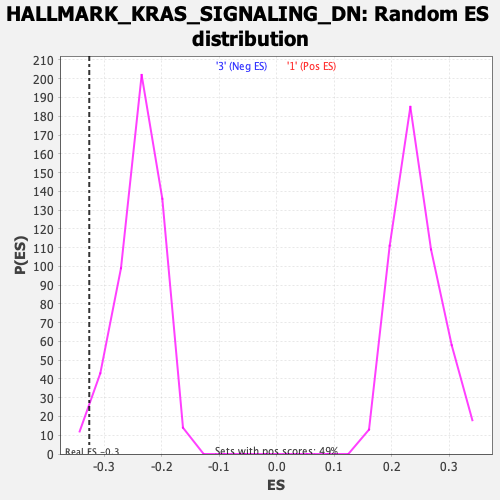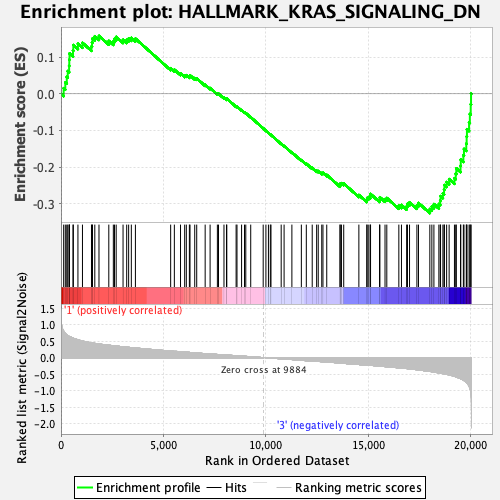
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | GMP.GMP.neu_Pheno.cls #Group2_versus_Group4.GMP.neu_Pheno.cls #Group2_versus_Group4_repos |
| Phenotype | GMP.neu_Pheno.cls#Group2_versus_Group4_repos |
| Upregulated in class | 3 |
| GeneSet | HALLMARK_KRAS_SIGNALING_DN |
| Enrichment Score (ES) | -0.32588872 |
| Normalized Enrichment Score (NES) | -1.3649558 |
| Nominal p-value | 0.019762846 |
| FDR q-value | 1.0 |
| FWER p-Value | 0.627 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Zfp112 | 130 | 0.804 | 0.0153 | No |
| 2 | Camk1d | 210 | 0.732 | 0.0312 | No |
| 3 | Sidt1 | 282 | 0.688 | 0.0464 | No |
| 4 | Dcc | 330 | 0.668 | 0.0621 | No |
| 5 | Prodh | 396 | 0.644 | 0.0763 | No |
| 6 | Kcnn1 | 405 | 0.642 | 0.0934 | No |
| 7 | Mefv | 415 | 0.639 | 0.1103 | No |
| 8 | Cacna1f | 585 | 0.597 | 0.1180 | No |
| 9 | Itgb1bp2 | 602 | 0.589 | 0.1332 | No |
| 10 | Tg | 826 | 0.541 | 0.1367 | No |
| 11 | Cpeb3 | 1047 | 0.502 | 0.1393 | No |
| 12 | Rgs11 | 1487 | 0.448 | 0.1294 | No |
| 13 | Ybx2 | 1513 | 0.445 | 0.1402 | No |
| 14 | Abcg4 | 1536 | 0.441 | 0.1511 | No |
| 15 | Hsd11b2 | 1649 | 0.431 | 0.1572 | No |
| 16 | Ngb | 1853 | 0.414 | 0.1582 | No |
| 17 | Atp4a | 2332 | 0.376 | 0.1445 | No |
| 18 | Tgm1 | 2553 | 0.359 | 0.1432 | No |
| 19 | Hnf1a | 2609 | 0.355 | 0.1501 | No |
| 20 | Sgk1 | 2692 | 0.350 | 0.1555 | No |
| 21 | Ryr2 | 3027 | 0.327 | 0.1476 | No |
| 22 | Slc12a3 | 3205 | 0.316 | 0.1473 | No |
| 23 | Gpr19 | 3300 | 0.312 | 0.1510 | No |
| 24 | Btg2 | 3431 | 0.304 | 0.1528 | No |
| 25 | Hc | 3630 | 0.292 | 0.1508 | No |
| 26 | P2rx6 | 5350 | 0.199 | 0.0699 | No |
| 27 | Clstn3 | 5530 | 0.190 | 0.0660 | No |
| 28 | Bard1 | 5831 | 0.178 | 0.0558 | No |
| 29 | Entpd7 | 6034 | 0.170 | 0.0503 | No |
| 30 | Klk8 | 6114 | 0.166 | 0.0508 | No |
| 31 | Nr6a1 | 6263 | 0.159 | 0.0477 | No |
| 32 | Nos1 | 6294 | 0.157 | 0.0505 | No |
| 33 | Htr1d | 6524 | 0.149 | 0.0430 | No |
| 34 | Ptprj | 6621 | 0.143 | 0.0421 | No |
| 35 | Slc5a5 | 7033 | 0.121 | 0.0248 | No |
| 36 | Abcb11 | 7274 | 0.110 | 0.0157 | No |
| 37 | Skil | 7629 | 0.097 | 0.0006 | No |
| 38 | Plag1 | 7684 | 0.094 | 0.0004 | No |
| 39 | Tex15 | 7951 | 0.086 | -0.0106 | No |
| 40 | Nrip2 | 8074 | 0.080 | -0.0146 | No |
| 41 | Vps50 | 8089 | 0.079 | -0.0131 | No |
| 42 | Lfng | 8543 | 0.060 | -0.0342 | No |
| 43 | Kcnd1 | 8586 | 0.058 | -0.0347 | No |
| 44 | Gtf3c5 | 8810 | 0.048 | -0.0446 | No |
| 45 | Selenop | 8953 | 0.042 | -0.0506 | No |
| 46 | Grid2 | 9015 | 0.039 | -0.0526 | No |
| 47 | Gp1ba | 9256 | 0.030 | -0.0639 | No |
| 48 | Cdkal1 | 9864 | 0.001 | -0.0943 | No |
| 49 | Tcf7l1 | 9997 | -0.002 | -0.1009 | No |
| 50 | Fggy | 10120 | -0.008 | -0.1068 | No |
| 51 | Epha5 | 10212 | -0.011 | -0.1111 | No |
| 52 | Arpp21 | 10244 | -0.012 | -0.1123 | No |
| 53 | Kmt2d | 10742 | -0.032 | -0.1364 | No |
| 54 | Prkn | 10884 | -0.038 | -0.1424 | No |
| 55 | Mthfr | 11264 | -0.055 | -0.1599 | No |
| 56 | Nr4a2 | 11726 | -0.077 | -0.1810 | No |
| 57 | Copz2 | 11968 | -0.089 | -0.1907 | No |
| 58 | Mfsd6 | 12256 | -0.102 | -0.2023 | No |
| 59 | Coq8a | 12472 | -0.110 | -0.2101 | No |
| 60 | Efhd1 | 12550 | -0.114 | -0.2109 | No |
| 61 | Cd80 | 12721 | -0.120 | -0.2162 | No |
| 62 | Asb7 | 12780 | -0.122 | -0.2158 | No |
| 63 | Tgfb2 | 12965 | -0.131 | -0.2215 | No |
| 64 | Edar | 13613 | -0.162 | -0.2496 | No |
| 65 | Bmpr1b | 13618 | -0.162 | -0.2454 | No |
| 66 | Stag3 | 13682 | -0.165 | -0.2440 | No |
| 67 | Tnni3 | 13793 | -0.171 | -0.2449 | No |
| 68 | Serpinb2 | 14532 | -0.207 | -0.2764 | No |
| 69 | Mast3 | 14917 | -0.225 | -0.2895 | No |
| 70 | Myo15a | 14936 | -0.226 | -0.2843 | No |
| 71 | Arhgdig | 15016 | -0.230 | -0.2820 | No |
| 72 | Slc6a3 | 15088 | -0.234 | -0.2792 | No |
| 73 | Kcnmb1 | 15102 | -0.235 | -0.2735 | No |
| 74 | Slc29a3 | 15539 | -0.253 | -0.2885 | No |
| 75 | Sphk2 | 15558 | -0.255 | -0.2825 | No |
| 76 | Msh5 | 15808 | -0.269 | -0.2877 | No |
| 77 | Synpo | 15895 | -0.273 | -0.2846 | No |
| 78 | Tshb | 16482 | -0.306 | -0.3057 | No |
| 79 | Snn | 16610 | -0.313 | -0.3036 | No |
| 80 | Pde6b | 16863 | -0.328 | -0.3073 | No |
| 81 | Tfcp2l1 | 16903 | -0.330 | -0.3003 | No |
| 82 | Macroh2a2 | 17004 | -0.338 | -0.2962 | No |
| 83 | Tent5c | 17363 | -0.363 | -0.3043 | No |
| 84 | Rsad2 | 17440 | -0.367 | -0.2981 | No |
| 85 | Fgf22 | 17994 | -0.415 | -0.3146 | Yes |
| 86 | Tenm2 | 18093 | -0.424 | -0.3080 | Yes |
| 87 | Cyp39a1 | 18193 | -0.435 | -0.3012 | Yes |
| 88 | Brdt | 18435 | -0.460 | -0.3008 | Yes |
| 89 | Zc2hc1c | 18501 | -0.468 | -0.2914 | Yes |
| 90 | Zbtb16 | 18517 | -0.470 | -0.2794 | Yes |
| 91 | Magix | 18632 | -0.481 | -0.2720 | Yes |
| 92 | Dtnb | 18689 | -0.486 | -0.2617 | Yes |
| 93 | Serpina10 | 18706 | -0.488 | -0.2492 | Yes |
| 94 | Cpa2 | 18814 | -0.501 | -0.2410 | Yes |
| 95 | Capn9 | 18936 | -0.519 | -0.2330 | Yes |
| 96 | Ccdc106 | 19195 | -0.565 | -0.2306 | Yes |
| 97 | Ypel1 | 19258 | -0.576 | -0.2181 | Yes |
| 98 | Thnsl2 | 19286 | -0.582 | -0.2036 | Yes |
| 99 | Gamt | 19492 | -0.633 | -0.1967 | Yes |
| 100 | Pdk2 | 19504 | -0.638 | -0.1799 | Yes |
| 101 | Idua | 19630 | -0.677 | -0.1678 | Yes |
| 102 | Egf | 19660 | -0.691 | -0.1505 | Yes |
| 103 | Sptbn2 | 19772 | -0.739 | -0.1360 | Yes |
| 104 | Thrb | 19791 | -0.748 | -0.1166 | Yes |
| 105 | Celsr2 | 19805 | -0.757 | -0.0967 | Yes |
| 106 | Ryr1 | 19909 | -0.868 | -0.0783 | Yes |
| 107 | Slc16a7 | 19947 | -0.925 | -0.0551 | Yes |
| 108 | Slc25a23 | 19989 | -1.052 | -0.0285 | Yes |
| 109 | Mx2 | 20005 | -1.117 | 0.0011 | Yes |