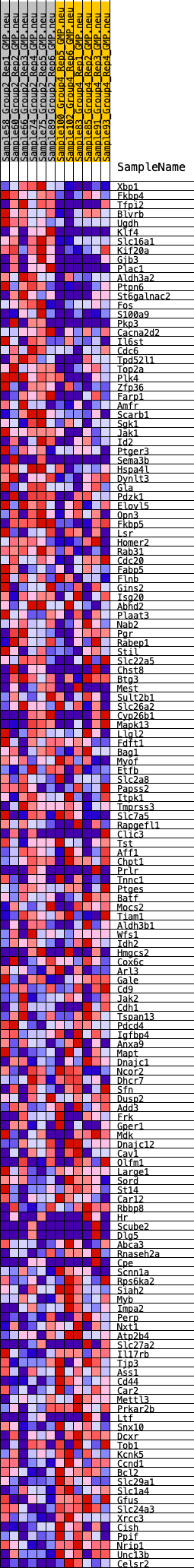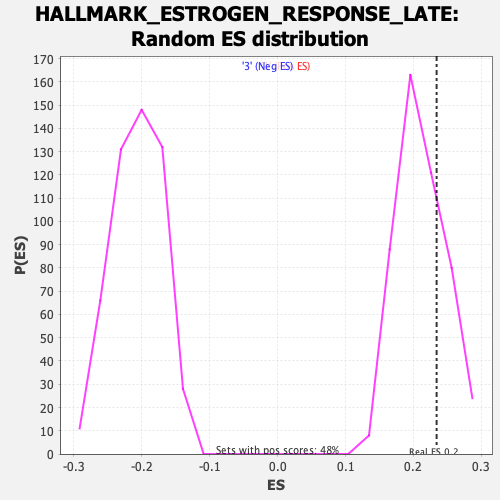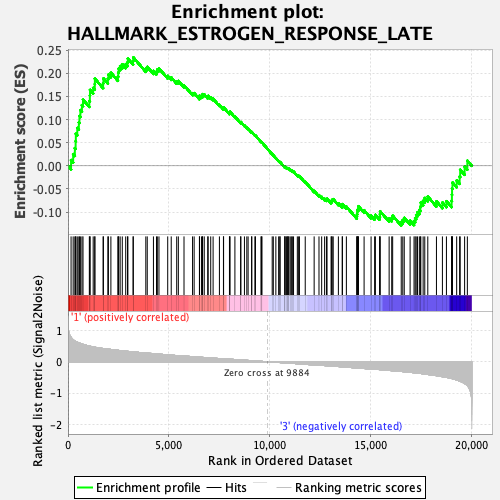
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | GMP.GMP.neu_Pheno.cls #Group2_versus_Group4.GMP.neu_Pheno.cls #Group2_versus_Group4_repos |
| Phenotype | GMP.neu_Pheno.cls#Group2_versus_Group4_repos |
| Upregulated in class | 1 |
| GeneSet | HALLMARK_ESTROGEN_RESPONSE_LATE |
| Enrichment Score (ES) | 0.23403618 |
| Normalized Enrichment Score (NES) | 1.1098456 |
| Nominal p-value | 0.2644628 |
| FDR q-value | 0.58223605 |
| FWER p-Value | 0.939 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Xbp1 | 152 | 0.781 | 0.0125 | Yes |
| 2 | Fkbp4 | 256 | 0.701 | 0.0253 | Yes |
| 3 | Tfpi2 | 340 | 0.665 | 0.0382 | Yes |
| 4 | Blvrb | 378 | 0.649 | 0.0531 | Yes |
| 5 | Ugdh | 389 | 0.646 | 0.0692 | Yes |
| 6 | Klf4 | 460 | 0.625 | 0.0818 | Yes |
| 7 | Slc16a1 | 538 | 0.608 | 0.0935 | Yes |
| 8 | Kif20a | 567 | 0.599 | 0.1076 | Yes |
| 9 | Gjb3 | 614 | 0.585 | 0.1203 | Yes |
| 10 | Plac1 | 691 | 0.568 | 0.1311 | Yes |
| 11 | Aldh3a2 | 744 | 0.557 | 0.1428 | Yes |
| 12 | Ptpn6 | 1058 | 0.501 | 0.1400 | Yes |
| 13 | St6galnac2 | 1085 | 0.498 | 0.1515 | Yes |
| 14 | Fos | 1092 | 0.496 | 0.1639 | Yes |
| 15 | S100a9 | 1249 | 0.475 | 0.1683 | Yes |
| 16 | Pkp3 | 1328 | 0.465 | 0.1764 | Yes |
| 17 | Cacna2d2 | 1333 | 0.464 | 0.1881 | Yes |
| 18 | Il6st | 1741 | 0.423 | 0.1785 | Yes |
| 19 | Cdc6 | 1756 | 0.422 | 0.1887 | Yes |
| 20 | Tpd52l1 | 1978 | 0.402 | 0.1879 | Yes |
| 21 | Top2a | 2003 | 0.401 | 0.1970 | Yes |
| 22 | Plk4 | 2127 | 0.394 | 0.2010 | Yes |
| 23 | Zfp36 | 2476 | 0.366 | 0.1929 | Yes |
| 24 | Farp1 | 2493 | 0.366 | 0.2015 | Yes |
| 25 | Amfr | 2513 | 0.364 | 0.2099 | Yes |
| 26 | Scarb1 | 2586 | 0.357 | 0.2155 | Yes |
| 27 | Sgk1 | 2692 | 0.350 | 0.2192 | Yes |
| 28 | Jak1 | 2862 | 0.339 | 0.2194 | Yes |
| 29 | Id2 | 2952 | 0.334 | 0.2235 | Yes |
| 30 | Ptger3 | 2969 | 0.333 | 0.2313 | Yes |
| 31 | Sema3b | 3227 | 0.315 | 0.2265 | Yes |
| 32 | Hspa4l | 3239 | 0.315 | 0.2340 | Yes |
| 33 | Dynlt3 | 3859 | 0.280 | 0.2101 | No |
| 34 | Gla | 3930 | 0.275 | 0.2136 | No |
| 35 | Pdzk1 | 4237 | 0.259 | 0.2049 | No |
| 36 | Elovl5 | 4390 | 0.250 | 0.2037 | No |
| 37 | Opn3 | 4429 | 0.247 | 0.2081 | No |
| 38 | Fkbp5 | 4511 | 0.242 | 0.2103 | No |
| 39 | Lsr | 4945 | 0.220 | 0.1942 | No |
| 40 | Homer2 | 5119 | 0.211 | 0.1909 | No |
| 41 | Rab31 | 5391 | 0.197 | 0.1823 | No |
| 42 | Cdc20 | 5472 | 0.192 | 0.1832 | No |
| 43 | Fabp5 | 5753 | 0.182 | 0.1738 | No |
| 44 | Flnb | 6177 | 0.163 | 0.1567 | No |
| 45 | Gins2 | 6256 | 0.159 | 0.1569 | No |
| 46 | Isg20 | 6519 | 0.149 | 0.1476 | No |
| 47 | Abhd2 | 6520 | 0.149 | 0.1514 | No |
| 48 | Plaat3 | 6618 | 0.143 | 0.1502 | No |
| 49 | Nab2 | 6671 | 0.140 | 0.1512 | No |
| 50 | Pgr | 6674 | 0.140 | 0.1547 | No |
| 51 | Rabep1 | 6757 | 0.136 | 0.1541 | No |
| 52 | Stil | 6931 | 0.126 | 0.1486 | No |
| 53 | Slc22a5 | 6949 | 0.126 | 0.1510 | No |
| 54 | Chst8 | 7078 | 0.120 | 0.1477 | No |
| 55 | Btg3 | 7196 | 0.114 | 0.1447 | No |
| 56 | Mest | 7509 | 0.102 | 0.1317 | No |
| 57 | Sult2b1 | 7709 | 0.094 | 0.1240 | No |
| 58 | Slc26a2 | 7716 | 0.093 | 0.1261 | No |
| 59 | Cyp26b1 | 8007 | 0.083 | 0.1137 | No |
| 60 | Mapk13 | 8022 | 0.082 | 0.1151 | No |
| 61 | Llgl2 | 8030 | 0.082 | 0.1168 | No |
| 62 | Fdft1 | 8277 | 0.071 | 0.1063 | No |
| 63 | Bag1 | 8562 | 0.059 | 0.0935 | No |
| 64 | Myof | 8572 | 0.059 | 0.0946 | No |
| 65 | Etfb | 8750 | 0.051 | 0.0870 | No |
| 66 | Slc2a8 | 8868 | 0.046 | 0.0823 | No |
| 67 | Papss2 | 8932 | 0.043 | 0.0802 | No |
| 68 | Itpk1 | 9106 | 0.035 | 0.0724 | No |
| 69 | Tmprss3 | 9115 | 0.035 | 0.0729 | No |
| 70 | Slc7a5 | 9269 | 0.029 | 0.0660 | No |
| 71 | Rapgefl1 | 9297 | 0.028 | 0.0654 | No |
| 72 | Clic3 | 9570 | 0.015 | 0.0520 | No |
| 73 | Tst | 9598 | 0.013 | 0.0510 | No |
| 74 | Aff1 | 9619 | 0.012 | 0.0503 | No |
| 75 | Chpt1 | 10127 | -0.008 | 0.0250 | No |
| 76 | Prlr | 10182 | -0.011 | 0.0226 | No |
| 77 | Tnnc1 | 10308 | -0.015 | 0.0167 | No |
| 78 | Ptges | 10446 | -0.021 | 0.0103 | No |
| 79 | Batf | 10504 | -0.023 | 0.0081 | No |
| 80 | Mocs2 | 10518 | -0.023 | 0.0080 | No |
| 81 | Tiam1 | 10756 | -0.033 | -0.0031 | No |
| 82 | Aldh3b1 | 10757 | -0.033 | -0.0022 | No |
| 83 | Wfs1 | 10761 | -0.033 | -0.0015 | No |
| 84 | Idh2 | 10842 | -0.036 | -0.0046 | No |
| 85 | Hmgcs2 | 10869 | -0.037 | -0.0050 | No |
| 86 | Cox6c | 10896 | -0.039 | -0.0053 | No |
| 87 | Arl3 | 10926 | -0.041 | -0.0057 | No |
| 88 | Gale | 10956 | -0.042 | -0.0061 | No |
| 89 | Cd9 | 11022 | -0.044 | -0.0082 | No |
| 90 | Jak2 | 11092 | -0.047 | -0.0105 | No |
| 91 | Cdh1 | 11125 | -0.048 | -0.0108 | No |
| 92 | Tspan13 | 11185 | -0.052 | -0.0125 | No |
| 93 | Pdcd4 | 11373 | -0.061 | -0.0203 | No |
| 94 | Igfbp4 | 11438 | -0.064 | -0.0219 | No |
| 95 | Anxa9 | 11477 | -0.065 | -0.0221 | No |
| 96 | Mapt | 11765 | -0.079 | -0.0346 | No |
| 97 | Dnajc1 | 12207 | -0.099 | -0.0542 | No |
| 98 | Ncor2 | 12447 | -0.110 | -0.0634 | No |
| 99 | Dhcr7 | 12580 | -0.114 | -0.0671 | No |
| 100 | Sfn | 12726 | -0.120 | -0.0713 | No |
| 101 | Dusp2 | 12817 | -0.123 | -0.0727 | No |
| 102 | Add3 | 12846 | -0.124 | -0.0709 | No |
| 103 | Frk | 13047 | -0.134 | -0.0775 | No |
| 104 | Gper1 | 13076 | -0.136 | -0.0754 | No |
| 105 | Mdk | 13100 | -0.137 | -0.0730 | No |
| 106 | Dnajc12 | 13152 | -0.140 | -0.0720 | No |
| 107 | Cav1 | 13409 | -0.150 | -0.0810 | No |
| 108 | Olfm1 | 13600 | -0.161 | -0.0865 | No |
| 109 | Large1 | 13609 | -0.161 | -0.0827 | No |
| 110 | Sord | 13805 | -0.171 | -0.0881 | No |
| 111 | St14 | 14324 | -0.196 | -0.1091 | No |
| 112 | Car12 | 14332 | -0.197 | -0.1044 | No |
| 113 | Rbbp8 | 14348 | -0.197 | -0.1001 | No |
| 114 | Hr | 14350 | -0.198 | -0.0951 | No |
| 115 | Scube2 | 14399 | -0.200 | -0.0923 | No |
| 116 | Dlg5 | 14402 | -0.200 | -0.0873 | No |
| 117 | Abca3 | 14684 | -0.214 | -0.0959 | No |
| 118 | Rnaseh2a | 15030 | -0.231 | -0.1073 | No |
| 119 | Cpe | 15206 | -0.237 | -0.1101 | No |
| 120 | Scnn1a | 15244 | -0.238 | -0.1058 | No |
| 121 | Rps6ka2 | 15452 | -0.249 | -0.1098 | No |
| 122 | Siah2 | 15470 | -0.249 | -0.1042 | No |
| 123 | Myb | 15478 | -0.250 | -0.0982 | No |
| 124 | Impa2 | 15926 | -0.275 | -0.1136 | No |
| 125 | Perp | 16054 | -0.282 | -0.1127 | No |
| 126 | Nxt1 | 16101 | -0.284 | -0.1077 | No |
| 127 | Atp2b4 | 16526 | -0.308 | -0.1211 | No |
| 128 | Slc27a2 | 16603 | -0.312 | -0.1169 | No |
| 129 | Il17rb | 16675 | -0.316 | -0.1123 | No |
| 130 | Tjp3 | 16966 | -0.335 | -0.1183 | No |
| 131 | Ass1 | 17161 | -0.348 | -0.1191 | No |
| 132 | Cd44 | 17217 | -0.353 | -0.1128 | No |
| 133 | Car2 | 17268 | -0.357 | -0.1061 | No |
| 134 | Mettl3 | 17334 | -0.361 | -0.1001 | No |
| 135 | Prkar2b | 17424 | -0.366 | -0.0951 | No |
| 136 | Ltf | 17475 | -0.371 | -0.0881 | No |
| 137 | Snx10 | 17497 | -0.373 | -0.0795 | No |
| 138 | Dcxr | 17612 | -0.383 | -0.0754 | No |
| 139 | Tob1 | 17693 | -0.390 | -0.0694 | No |
| 140 | Kcnk5 | 17843 | -0.405 | -0.0665 | No |
| 141 | Ccnd1 | 18272 | -0.442 | -0.0766 | No |
| 142 | Bcl2 | 18568 | -0.476 | -0.0792 | No |
| 143 | Slc29a1 | 18767 | -0.495 | -0.0765 | No |
| 144 | Slc1a4 | 19023 | -0.533 | -0.0756 | No |
| 145 | Gfus | 19039 | -0.536 | -0.0625 | No |
| 146 | Slc24a3 | 19047 | -0.537 | -0.0491 | No |
| 147 | Xrcc3 | 19067 | -0.539 | -0.0361 | No |
| 148 | Cish | 19275 | -0.579 | -0.0317 | No |
| 149 | Ppif | 19422 | -0.617 | -0.0231 | No |
| 150 | Nrip1 | 19449 | -0.624 | -0.0084 | No |
| 151 | Unc13b | 19669 | -0.693 | -0.0016 | No |
| 152 | Celsr2 | 19805 | -0.757 | 0.0111 | No |