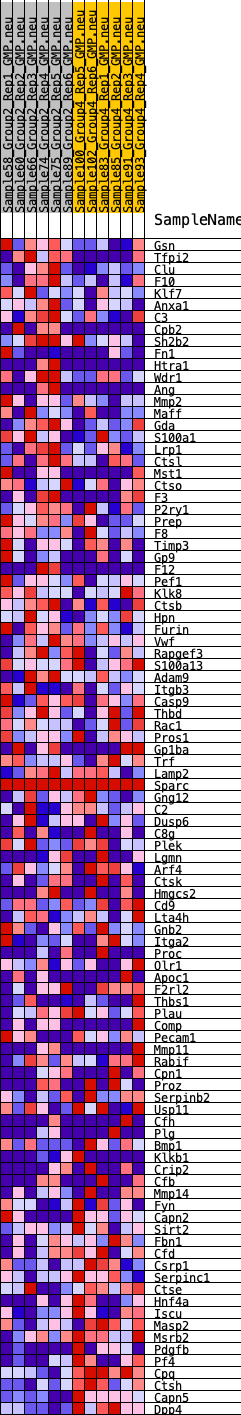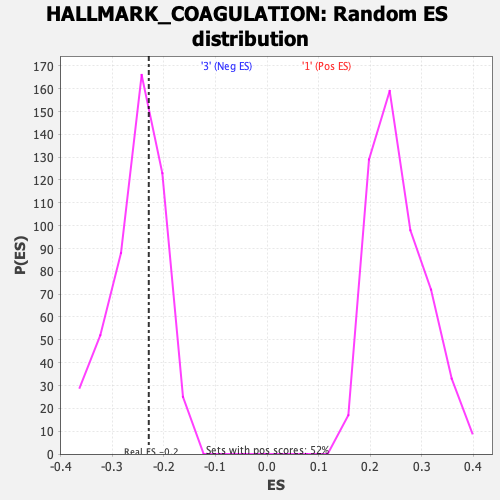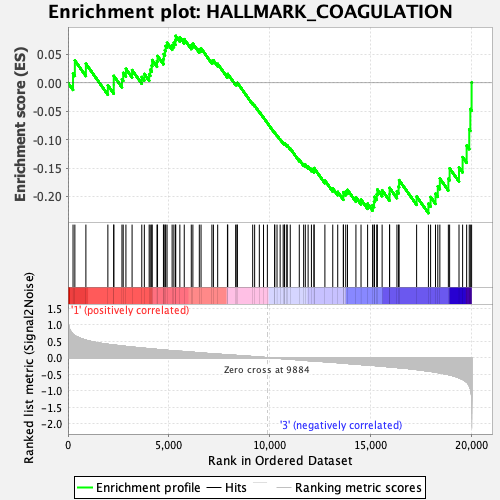
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | GMP.GMP.neu_Pheno.cls #Group2_versus_Group4.GMP.neu_Pheno.cls #Group2_versus_Group4_repos |
| Phenotype | GMP.neu_Pheno.cls#Group2_versus_Group4_repos |
| Upregulated in class | 3 |
| GeneSet | HALLMARK_COAGULATION |
| Enrichment Score (ES) | -0.22941041 |
| Normalized Enrichment Score (NES) | -0.90925753 |
| Nominal p-value | 0.6231884 |
| FDR q-value | 0.9587335 |
| FWER p-Value | 0.995 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Gsn | 250 | 0.705 | 0.0164 | No |
| 2 | Tfpi2 | 340 | 0.665 | 0.0393 | No |
| 3 | Clu | 887 | 0.530 | 0.0336 | No |
| 4 | F10 | 1977 | 0.402 | -0.0045 | No |
| 5 | Klf7 | 2269 | 0.381 | -0.0034 | No |
| 6 | Anxa1 | 2270 | 0.381 | 0.0122 | No |
| 7 | C3 | 2676 | 0.351 | 0.0063 | No |
| 8 | Cpb2 | 2737 | 0.347 | 0.0175 | No |
| 9 | Sh2b2 | 2872 | 0.339 | 0.0247 | No |
| 10 | Fn1 | 3178 | 0.317 | 0.0225 | No |
| 11 | Htra1 | 3659 | 0.291 | 0.0103 | No |
| 12 | Wdr1 | 3786 | 0.283 | 0.0156 | No |
| 13 | Ang | 4025 | 0.270 | 0.0148 | No |
| 14 | Mmp2 | 4078 | 0.268 | 0.0231 | No |
| 15 | Maff | 4151 | 0.265 | 0.0304 | No |
| 16 | Gda | 4175 | 0.264 | 0.0401 | No |
| 17 | S100a1 | 4414 | 0.248 | 0.0384 | No |
| 18 | Lrp1 | 4440 | 0.246 | 0.0472 | No |
| 19 | Ctsl | 4733 | 0.232 | 0.0421 | No |
| 20 | Mst1 | 4751 | 0.231 | 0.0508 | No |
| 21 | Ctso | 4807 | 0.228 | 0.0574 | No |
| 22 | F3 | 4843 | 0.226 | 0.0649 | No |
| 23 | P2ry1 | 4912 | 0.222 | 0.0706 | No |
| 24 | Prep | 5164 | 0.209 | 0.0666 | No |
| 25 | F8 | 5251 | 0.206 | 0.0707 | No |
| 26 | Timp3 | 5329 | 0.201 | 0.0751 | No |
| 27 | Gp9 | 5343 | 0.200 | 0.0826 | No |
| 28 | F12 | 5547 | 0.189 | 0.0802 | No |
| 29 | Pef1 | 5765 | 0.182 | 0.0768 | No |
| 30 | Klk8 | 6114 | 0.166 | 0.0662 | No |
| 31 | Ctsb | 6193 | 0.163 | 0.0689 | No |
| 32 | Hpn | 6512 | 0.150 | 0.0591 | No |
| 33 | Furin | 6598 | 0.145 | 0.0608 | No |
| 34 | Vwf | 7136 | 0.117 | 0.0387 | No |
| 35 | Rapgef3 | 7211 | 0.114 | 0.0396 | No |
| 36 | S100a13 | 7423 | 0.106 | 0.0334 | No |
| 37 | Adam9 | 7907 | 0.088 | 0.0127 | No |
| 38 | Itgb3 | 7922 | 0.087 | 0.0156 | No |
| 39 | Casp9 | 8314 | 0.069 | -0.0012 | No |
| 40 | Thbd | 8376 | 0.066 | -0.0015 | No |
| 41 | Rac1 | 8400 | 0.065 | 0.0000 | No |
| 42 | Pros1 | 9159 | 0.033 | -0.0366 | No |
| 43 | Gp1ba | 9256 | 0.030 | -0.0402 | No |
| 44 | Trf | 9488 | 0.018 | -0.0511 | No |
| 45 | Lamp2 | 9696 | 0.009 | -0.0611 | No |
| 46 | Sparc | 9887 | 0.000 | -0.0706 | No |
| 47 | Gng12 | 10249 | -0.013 | -0.0882 | No |
| 48 | C2 | 10251 | -0.013 | -0.0878 | No |
| 49 | Dusp6 | 10366 | -0.017 | -0.0928 | No |
| 50 | C8g | 10521 | -0.024 | -0.0995 | No |
| 51 | Plek | 10671 | -0.030 | -0.1058 | No |
| 52 | Lgmn | 10734 | -0.032 | -0.1076 | No |
| 53 | Arf4 | 10743 | -0.032 | -0.1067 | No |
| 54 | Ctsk | 10852 | -0.037 | -0.1106 | No |
| 55 | Hmgcs2 | 10869 | -0.037 | -0.1098 | No |
| 56 | Cd9 | 11022 | -0.044 | -0.1156 | No |
| 57 | Lta4h | 11470 | -0.065 | -0.1354 | No |
| 58 | Gnb2 | 11686 | -0.075 | -0.1431 | No |
| 59 | Itga2 | 11769 | -0.079 | -0.1440 | No |
| 60 | Proc | 11908 | -0.087 | -0.1473 | No |
| 61 | Olr1 | 12072 | -0.094 | -0.1517 | No |
| 62 | Apoc1 | 12201 | -0.099 | -0.1540 | No |
| 63 | F2rl2 | 12203 | -0.099 | -0.1500 | No |
| 64 | Thbs1 | 12740 | -0.120 | -0.1720 | No |
| 65 | Plau | 13130 | -0.139 | -0.1858 | No |
| 66 | Comp | 13376 | -0.149 | -0.1920 | No |
| 67 | Pecam1 | 13656 | -0.164 | -0.1992 | No |
| 68 | Mmp11 | 13658 | -0.164 | -0.1925 | No |
| 69 | Rabif | 13773 | -0.170 | -0.1913 | No |
| 70 | Cpn1 | 13860 | -0.174 | -0.1884 | No |
| 71 | Proz | 14279 | -0.194 | -0.2014 | No |
| 72 | Serpinb2 | 14532 | -0.207 | -0.2056 | No |
| 73 | Usp11 | 14858 | -0.223 | -0.2127 | No |
| 74 | Cfh | 15105 | -0.235 | -0.2154 | Yes |
| 75 | Plg | 15178 | -0.235 | -0.2094 | Yes |
| 76 | Bmp1 | 15199 | -0.237 | -0.2007 | Yes |
| 77 | Klkb1 | 15312 | -0.242 | -0.1963 | Yes |
| 78 | Crip2 | 15341 | -0.243 | -0.1877 | Yes |
| 79 | Cfb | 15578 | -0.256 | -0.1891 | Yes |
| 80 | Mmp14 | 15946 | -0.276 | -0.1962 | Yes |
| 81 | Fyn | 15950 | -0.276 | -0.1850 | Yes |
| 82 | Capn2 | 16313 | -0.297 | -0.1909 | Yes |
| 83 | Sirt2 | 16394 | -0.302 | -0.1825 | Yes |
| 84 | Fbn1 | 16420 | -0.304 | -0.1713 | Yes |
| 85 | Cfd | 17289 | -0.359 | -0.2001 | Yes |
| 86 | Csrp1 | 17874 | -0.408 | -0.2126 | Yes |
| 87 | Serpinc1 | 17982 | -0.415 | -0.2010 | Yes |
| 88 | Ctse | 18225 | -0.438 | -0.1951 | Yes |
| 89 | Hnf4a | 18339 | -0.449 | -0.1823 | Yes |
| 90 | Iscu | 18441 | -0.460 | -0.1685 | Yes |
| 91 | Masp2 | 18863 | -0.509 | -0.1687 | Yes |
| 92 | Msrb2 | 18926 | -0.517 | -0.1506 | Yes |
| 93 | Pdgfb | 19391 | -0.610 | -0.1488 | Yes |
| 94 | Pf4 | 19570 | -0.656 | -0.1308 | Yes |
| 95 | Cpq | 19770 | -0.739 | -0.1104 | Yes |
| 96 | Ctsh | 19899 | -0.851 | -0.0819 | Yes |
| 97 | Capn5 | 19953 | -0.936 | -0.0461 | Yes |
| 98 | Dpp4 | 20016 | -1.210 | 0.0005 | Yes |