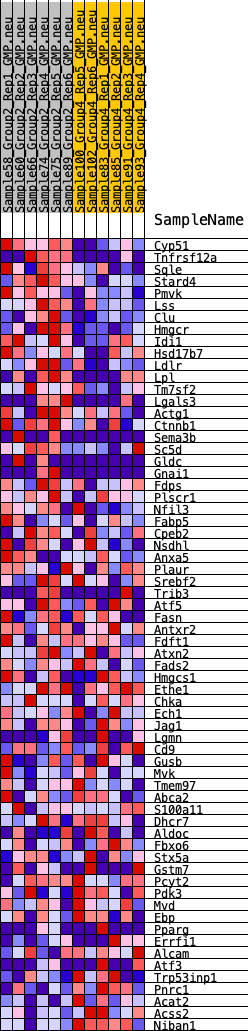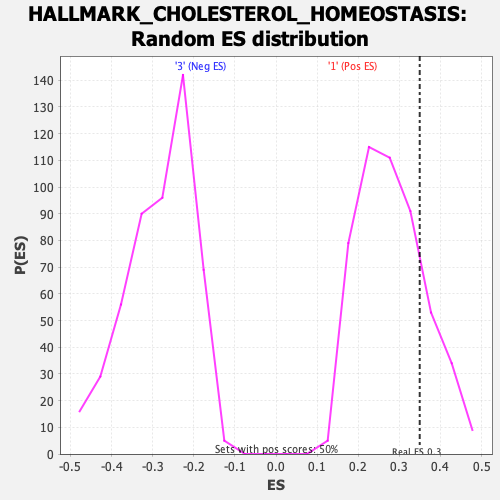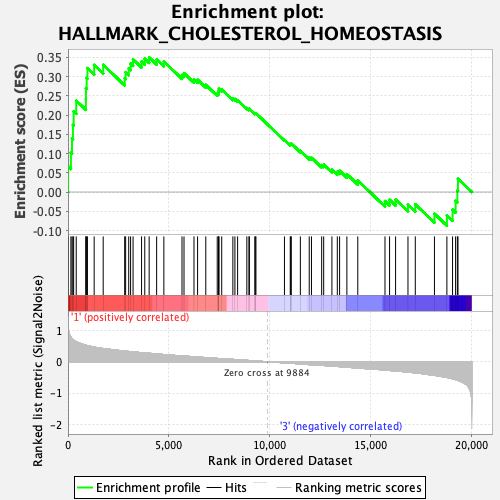
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | GMP.GMP.neu_Pheno.cls #Group2_versus_Group4.GMP.neu_Pheno.cls #Group2_versus_Group4_repos |
| Phenotype | GMP.neu_Pheno.cls#Group2_versus_Group4_repos |
| Upregulated in class | 1 |
| GeneSet | HALLMARK_CHOLESTEROL_HOMEOSTASIS |
| Enrichment Score (ES) | 0.3495715 |
| Normalized Enrichment Score (NES) | 1.2507417 |
| Nominal p-value | 0.1971831 |
| FDR q-value | 0.44938105 |
| FWER p-Value | 0.826 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Cyp51 | 7 | 1.253 | 0.0665 | Yes |
| 2 | Tnfrsf12a | 143 | 0.791 | 0.1020 | Yes |
| 3 | Sqle | 198 | 0.741 | 0.1388 | Yes |
| 4 | Stard4 | 245 | 0.708 | 0.1743 | Yes |
| 5 | Pmvk | 280 | 0.690 | 0.2094 | Yes |
| 6 | Lss | 408 | 0.640 | 0.2372 | Yes |
| 7 | Clu | 887 | 0.530 | 0.2416 | Yes |
| 8 | Hmgcr | 890 | 0.530 | 0.2698 | Yes |
| 9 | Idi1 | 925 | 0.523 | 0.2960 | Yes |
| 10 | Hsd17b7 | 963 | 0.517 | 0.3217 | Yes |
| 11 | Ldlr | 1297 | 0.468 | 0.3301 | Yes |
| 12 | Lpl | 1745 | 0.423 | 0.3302 | Yes |
| 13 | Tm7sf2 | 2811 | 0.344 | 0.2952 | Yes |
| 14 | Lgals3 | 2858 | 0.340 | 0.3111 | Yes |
| 15 | Actg1 | 3011 | 0.330 | 0.3210 | Yes |
| 16 | Ctnnb1 | 3104 | 0.323 | 0.3337 | Yes |
| 17 | Sema3b | 3227 | 0.315 | 0.3444 | Yes |
| 18 | Sc5d | 3648 | 0.291 | 0.3389 | Yes |
| 19 | Gldc | 3807 | 0.281 | 0.3460 | Yes |
| 20 | Gnai1 | 4024 | 0.270 | 0.3496 | Yes |
| 21 | Fdps | 4396 | 0.250 | 0.3443 | No |
| 22 | Plscr1 | 4754 | 0.231 | 0.3388 | No |
| 23 | Nfil3 | 5653 | 0.189 | 0.3038 | No |
| 24 | Fabp5 | 5753 | 0.182 | 0.3086 | No |
| 25 | Cpeb2 | 6248 | 0.160 | 0.2924 | No |
| 26 | Nsdhl | 6426 | 0.151 | 0.2916 | No |
| 27 | Anxa5 | 6833 | 0.131 | 0.2782 | No |
| 28 | Plaur | 7401 | 0.107 | 0.2555 | No |
| 29 | Srebf2 | 7463 | 0.104 | 0.2580 | No |
| 30 | Trib3 | 7474 | 0.104 | 0.2631 | No |
| 31 | Atf5 | 7486 | 0.103 | 0.2680 | No |
| 32 | Fasn | 7623 | 0.097 | 0.2664 | No |
| 33 | Antxr2 | 8176 | 0.076 | 0.2428 | No |
| 34 | Fdft1 | 8277 | 0.071 | 0.2416 | No |
| 35 | Atxn2 | 8411 | 0.065 | 0.2384 | No |
| 36 | Fads2 | 8869 | 0.046 | 0.2179 | No |
| 37 | Hmgcs1 | 8974 | 0.041 | 0.2149 | No |
| 38 | Ethe1 | 8994 | 0.040 | 0.2161 | No |
| 39 | Chka | 9268 | 0.029 | 0.2040 | No |
| 40 | Ech1 | 9285 | 0.028 | 0.2047 | No |
| 41 | Jag1 | 9320 | 0.027 | 0.2044 | No |
| 42 | Lgmn | 10734 | -0.032 | 0.1353 | No |
| 43 | Cd9 | 11022 | -0.044 | 0.1233 | No |
| 44 | Gusb | 11045 | -0.045 | 0.1246 | No |
| 45 | Mvk | 11072 | -0.046 | 0.1258 | No |
| 46 | Tmem97 | 11523 | -0.067 | 0.1068 | No |
| 47 | Abca2 | 11963 | -0.089 | 0.0896 | No |
| 48 | S100a11 | 12074 | -0.094 | 0.0891 | No |
| 49 | Dhcr7 | 12580 | -0.114 | 0.0698 | No |
| 50 | Aldoc | 12680 | -0.118 | 0.0712 | No |
| 51 | Fbxo6 | 13088 | -0.136 | 0.0581 | No |
| 52 | Stx5a | 13356 | -0.148 | 0.0526 | No |
| 53 | Gstm7 | 13469 | -0.154 | 0.0552 | No |
| 54 | Pcyt2 | 13830 | -0.173 | 0.0464 | No |
| 55 | Pdk3 | 14371 | -0.199 | 0.0299 | No |
| 56 | Mvd | 15720 | -0.263 | -0.0236 | No |
| 57 | Ebp | 15947 | -0.276 | -0.0201 | No |
| 58 | Pparg | 16246 | -0.293 | -0.0194 | No |
| 59 | Errfi1 | 16860 | -0.328 | -0.0326 | No |
| 60 | Alcam | 17223 | -0.353 | -0.0319 | No |
| 61 | Atf3 | 18173 | -0.432 | -0.0564 | No |
| 62 | Trp53inp1 | 18790 | -0.497 | -0.0607 | No |
| 63 | Pnrc1 | 19071 | -0.541 | -0.0459 | No |
| 64 | Acat2 | 19222 | -0.569 | -0.0230 | No |
| 65 | Acss2 | 19309 | -0.588 | 0.0041 | No |
| 66 | Niban1 | 19340 | -0.596 | 0.0344 | No |