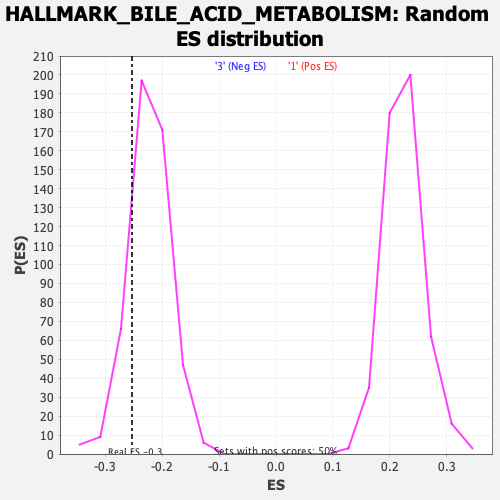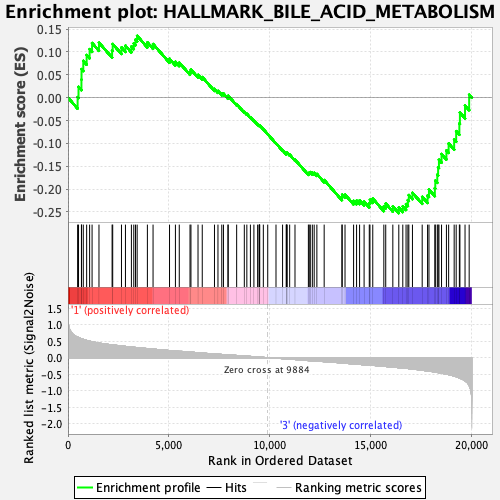
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | GMP.GMP.neu_Pheno.cls #Group2_versus_Group4.GMP.neu_Pheno.cls #Group2_versus_Group4_repos |
| Phenotype | GMP.neu_Pheno.cls#Group2_versus_Group4_repos |
| Upregulated in class | 3 |
| GeneSet | HALLMARK_BILE_ACID_METABOLISM |
| Enrichment Score (ES) | -0.25282493 |
| Normalized Enrichment Score (NES) | -1.1315895 |
| Nominal p-value | 0.17764471 |
| FDR q-value | 0.99953175 |
| FWER p-Value | 0.91 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Gclm | 477 | 0.622 | 0.0012 | No |
| 2 | Abca5 | 522 | 0.611 | 0.0236 | No |
| 3 | Dhcr24 | 666 | 0.574 | 0.0396 | No |
| 4 | Cyp46a1 | 671 | 0.573 | 0.0625 | No |
| 5 | Scp2 | 758 | 0.555 | 0.0806 | No |
| 6 | Idi1 | 925 | 0.523 | 0.0934 | No |
| 7 | Ephx2 | 1072 | 0.500 | 0.1062 | No |
| 8 | Klf1 | 1194 | 0.483 | 0.1196 | No |
| 9 | Abcg4 | 1536 | 0.441 | 0.1203 | No |
| 10 | Pecr | 2187 | 0.388 | 0.1034 | No |
| 11 | Lck | 2217 | 0.385 | 0.1174 | No |
| 12 | Slc27a5 | 2652 | 0.353 | 0.1099 | No |
| 13 | Efhc1 | 2849 | 0.341 | 0.1138 | No |
| 14 | Dio2 | 3144 | 0.320 | 0.1120 | No |
| 15 | Aldh9a1 | 3262 | 0.314 | 0.1188 | No |
| 16 | Hao1 | 3346 | 0.310 | 0.1271 | No |
| 17 | Idh1 | 3429 | 0.304 | 0.1353 | No |
| 18 | Gnpat | 3937 | 0.275 | 0.1209 | No |
| 19 | Fads1 | 4218 | 0.261 | 0.1174 | No |
| 20 | Abcd1 | 5035 | 0.215 | 0.0851 | No |
| 21 | Acsl5 | 5327 | 0.201 | 0.0786 | No |
| 22 | Aldh8a1 | 5518 | 0.190 | 0.0767 | No |
| 23 | Paox | 6051 | 0.169 | 0.0569 | No |
| 24 | Npc1 | 6097 | 0.167 | 0.0614 | No |
| 25 | Retsat | 6448 | 0.150 | 0.0498 | No |
| 26 | Acsl1 | 6658 | 0.141 | 0.0451 | No |
| 27 | Slc23a2 | 7264 | 0.110 | 0.0191 | No |
| 28 | Abcd3 | 7437 | 0.105 | 0.0147 | No |
| 29 | Fdxr | 7626 | 0.097 | 0.0092 | No |
| 30 | Sult2b1 | 7709 | 0.094 | 0.0089 | No |
| 31 | Amacr | 7934 | 0.087 | 0.0012 | No |
| 32 | Aqp9 | 7943 | 0.086 | 0.0042 | No |
| 33 | Lonp2 | 8364 | 0.067 | -0.0141 | No |
| 34 | Bcar3 | 8744 | 0.051 | -0.0311 | No |
| 35 | Fads2 | 8869 | 0.046 | -0.0355 | No |
| 36 | Cyp27a1 | 9046 | 0.038 | -0.0428 | No |
| 37 | Hsd17b4 | 9219 | 0.031 | -0.0502 | No |
| 38 | Rbp1 | 9404 | 0.023 | -0.0585 | No |
| 39 | Hacl1 | 9472 | 0.019 | -0.0611 | No |
| 40 | Ar | 9497 | 0.018 | -0.0615 | No |
| 41 | Sod1 | 9501 | 0.018 | -0.0610 | No |
| 42 | Pex6 | 9505 | 0.018 | -0.0604 | No |
| 43 | Pex11g | 9687 | 0.009 | -0.0691 | No |
| 44 | Gc | 9905 | 0.000 | -0.0800 | No |
| 45 | Pex11a | 10315 | -0.015 | -0.0999 | No |
| 46 | Pex16 | 10641 | -0.029 | -0.1151 | No |
| 47 | Soat2 | 10821 | -0.035 | -0.1226 | No |
| 48 | Idh2 | 10842 | -0.036 | -0.1221 | No |
| 49 | Crot | 10845 | -0.037 | -0.1208 | No |
| 50 | Pnpla8 | 10866 | -0.037 | -0.1203 | No |
| 51 | Bmp6 | 10993 | -0.043 | -0.1248 | No |
| 52 | Nedd4 | 11257 | -0.055 | -0.1358 | No |
| 53 | Pxmp2 | 11921 | -0.087 | -0.1656 | No |
| 54 | Abca2 | 11963 | -0.089 | -0.1640 | No |
| 55 | Optn | 12016 | -0.091 | -0.1630 | No |
| 56 | Pex26 | 12117 | -0.096 | -0.1641 | No |
| 57 | Rxra | 12209 | -0.100 | -0.1647 | No |
| 58 | Phyh | 12342 | -0.105 | -0.1671 | No |
| 59 | Slc23a1 | 12704 | -0.119 | -0.1804 | No |
| 60 | Aldh1a1 | 13584 | -0.160 | -0.2180 | No |
| 61 | Pex7 | 13598 | -0.161 | -0.2122 | No |
| 62 | Isoc1 | 13738 | -0.168 | -0.2124 | No |
| 63 | Pfkm | 14163 | -0.189 | -0.2261 | No |
| 64 | Slc22a18 | 14311 | -0.196 | -0.2255 | No |
| 65 | Pex1 | 14461 | -0.202 | -0.2249 | No |
| 66 | Abca3 | 14684 | -0.214 | -0.2274 | No |
| 67 | Abca6 | 14941 | -0.226 | -0.2311 | No |
| 68 | Gstk1 | 14975 | -0.228 | -0.2235 | No |
| 69 | Akr1d1 | 15114 | -0.235 | -0.2210 | No |
| 70 | Hsd17b11 | 15663 | -0.261 | -0.2380 | No |
| 71 | Cat | 15757 | -0.266 | -0.2319 | No |
| 72 | Slc35b2 | 16109 | -0.285 | -0.2380 | No |
| 73 | Abcd2 | 16405 | -0.303 | -0.2406 | Yes |
| 74 | Slc27a2 | 16603 | -0.312 | -0.2379 | Yes |
| 75 | Nr1h4 | 16765 | -0.322 | -0.2330 | Yes |
| 76 | Atxn1 | 16856 | -0.328 | -0.2243 | Yes |
| 77 | Tfcp2l1 | 16903 | -0.330 | -0.2132 | Yes |
| 78 | Nr0b2 | 17082 | -0.343 | -0.2083 | Yes |
| 79 | Prdx5 | 17565 | -0.379 | -0.2172 | Yes |
| 80 | Abca4 | 17832 | -0.405 | -0.2142 | Yes |
| 81 | Mlycd | 17901 | -0.409 | -0.2011 | Yes |
| 82 | Cyp39a1 | 18193 | -0.435 | -0.1982 | Yes |
| 83 | Lipe | 18215 | -0.437 | -0.1816 | Yes |
| 84 | Pex12 | 18314 | -0.447 | -0.1685 | Yes |
| 85 | Pex13 | 18355 | -0.450 | -0.1523 | Yes |
| 86 | Abca8b | 18396 | -0.455 | -0.1360 | Yes |
| 87 | Gnmt | 18525 | -0.471 | -0.1234 | Yes |
| 88 | Slc29a1 | 18767 | -0.495 | -0.1155 | Yes |
| 89 | Nr3c2 | 18877 | -0.512 | -0.1003 | Yes |
| 90 | Cyp7b1 | 19151 | -0.557 | -0.0916 | Yes |
| 91 | Abca1 | 19251 | -0.575 | -0.0734 | Yes |
| 92 | Hsd3b7 | 19409 | -0.614 | -0.0565 | Yes |
| 93 | Pex19 | 19436 | -0.620 | -0.0328 | Yes |
| 94 | Abca9 | 19690 | -0.700 | -0.0172 | Yes |
| 95 | Nudt12 | 19895 | -0.844 | 0.0066 | Yes |