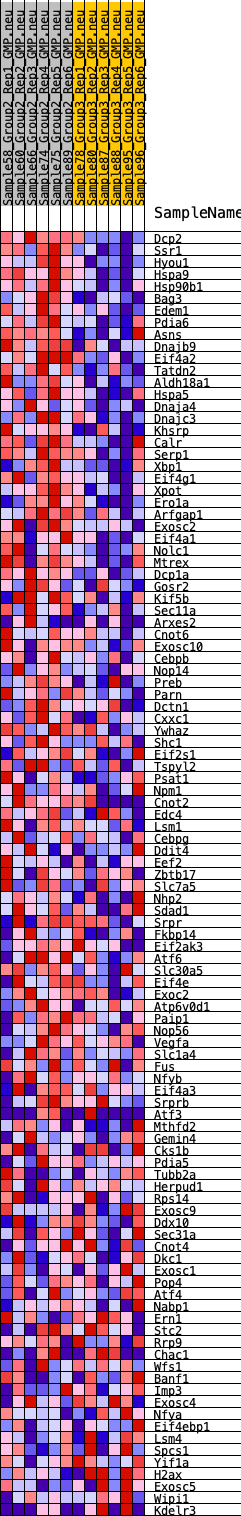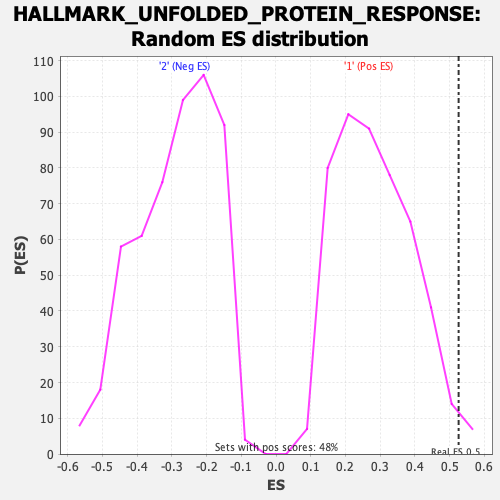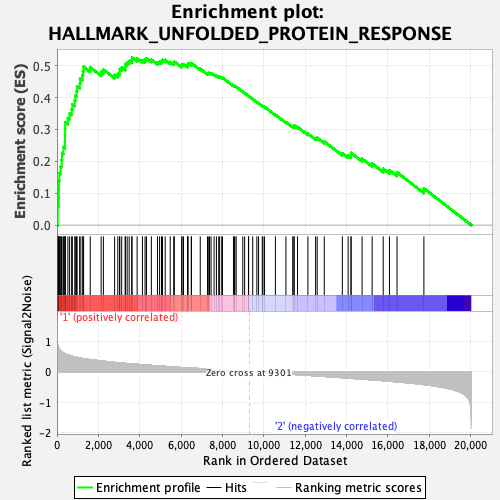
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | GMP.GMP.neu_Pheno.cls #Group2_versus_Group3.GMP.neu_Pheno.cls #Group2_versus_Group3_repos |
| Phenotype | GMP.neu_Pheno.cls#Group2_versus_Group3_repos |
| Upregulated in class | 1 |
| GeneSet | HALLMARK_UNFOLDED_PROTEIN_RESPONSE |
| Enrichment Score (ES) | 0.5258462 |
| Normalized Enrichment Score (NES) | 1.8418564 |
| Nominal p-value | 0.014644352 |
| FDR q-value | 0.023192903 |
| FWER p-Value | 0.035 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Dcp2 | 34 | 0.873 | 0.0290 | Yes |
| 2 | Ssr1 | 36 | 0.871 | 0.0595 | Yes |
| 3 | Hyou1 | 79 | 0.787 | 0.0850 | Yes |
| 4 | Hspa9 | 88 | 0.771 | 0.1117 | Yes |
| 5 | Hsp90b1 | 94 | 0.763 | 0.1383 | Yes |
| 6 | Bag3 | 115 | 0.730 | 0.1629 | Yes |
| 7 | Edem1 | 175 | 0.676 | 0.1837 | Yes |
| 8 | Pdia6 | 221 | 0.655 | 0.2044 | Yes |
| 9 | Asns | 242 | 0.646 | 0.2261 | Yes |
| 10 | Dnajb9 | 311 | 0.614 | 0.2443 | Yes |
| 11 | Eif4a2 | 383 | 0.590 | 0.2615 | Yes |
| 12 | Tatdn2 | 388 | 0.588 | 0.2819 | Yes |
| 13 | Aldh18a1 | 391 | 0.586 | 0.3024 | Yes |
| 14 | Hspa5 | 395 | 0.586 | 0.3228 | Yes |
| 15 | Dnaja4 | 527 | 0.552 | 0.3356 | Yes |
| 16 | Dnajc3 | 610 | 0.530 | 0.3501 | Yes |
| 17 | Khsrp | 708 | 0.509 | 0.3631 | Yes |
| 18 | Calr | 741 | 0.503 | 0.3792 | Yes |
| 19 | Serp1 | 863 | 0.485 | 0.3901 | Yes |
| 20 | Xbp1 | 883 | 0.482 | 0.4061 | Yes |
| 21 | Eif4g1 | 944 | 0.473 | 0.4197 | Yes |
| 22 | Xpot | 970 | 0.469 | 0.4350 | Yes |
| 23 | Ero1a | 1099 | 0.452 | 0.4444 | Yes |
| 24 | Arfgap1 | 1116 | 0.449 | 0.4594 | Yes |
| 25 | Exosc2 | 1219 | 0.437 | 0.4696 | Yes |
| 26 | Eif4a1 | 1258 | 0.432 | 0.4829 | Yes |
| 27 | Nolc1 | 1286 | 0.429 | 0.4966 | Yes |
| 28 | Mtrex | 1607 | 0.398 | 0.4945 | Yes |
| 29 | Dcp1a | 2134 | 0.354 | 0.4805 | Yes |
| 30 | Gosr2 | 2242 | 0.346 | 0.4873 | Yes |
| 31 | Kif5b | 2785 | 0.305 | 0.4708 | Yes |
| 32 | Sec11a | 2938 | 0.295 | 0.4736 | Yes |
| 33 | Arxes2 | 3025 | 0.290 | 0.4794 | Yes |
| 34 | Cnot6 | 3034 | 0.290 | 0.4892 | Yes |
| 35 | Exosc10 | 3127 | 0.284 | 0.4946 | Yes |
| 36 | Cebpb | 3300 | 0.274 | 0.4956 | Yes |
| 37 | Nop14 | 3302 | 0.274 | 0.5051 | Yes |
| 38 | Preb | 3392 | 0.269 | 0.5101 | Yes |
| 39 | Parn | 3488 | 0.265 | 0.5146 | Yes |
| 40 | Dctn1 | 3611 | 0.258 | 0.5176 | Yes |
| 41 | Cxxc1 | 3627 | 0.257 | 0.5258 | Yes |
| 42 | Ywhaz | 3873 | 0.243 | 0.5221 | No |
| 43 | Shc1 | 4130 | 0.230 | 0.5173 | No |
| 44 | Eif2s1 | 4257 | 0.224 | 0.5189 | No |
| 45 | Tspyl2 | 4325 | 0.219 | 0.5232 | No |
| 46 | Psat1 | 4561 | 0.208 | 0.5187 | No |
| 47 | Npm1 | 4856 | 0.191 | 0.5106 | No |
| 48 | Cnot2 | 4959 | 0.189 | 0.5122 | No |
| 49 | Edc4 | 5052 | 0.185 | 0.5140 | No |
| 50 | Lsm1 | 5100 | 0.182 | 0.5181 | No |
| 51 | Cebpg | 5230 | 0.176 | 0.5178 | No |
| 52 | Ddit4 | 5473 | 0.164 | 0.5114 | No |
| 53 | Eef2 | 5643 | 0.157 | 0.5084 | No |
| 54 | Zbtb17 | 5671 | 0.155 | 0.5125 | No |
| 55 | Slc7a5 | 6026 | 0.139 | 0.4996 | No |
| 56 | Nhp2 | 6046 | 0.138 | 0.5035 | No |
| 57 | Sdad1 | 6116 | 0.135 | 0.5048 | No |
| 58 | Srpr | 6310 | 0.128 | 0.4996 | No |
| 59 | Fkbp14 | 6313 | 0.128 | 0.5040 | No |
| 60 | Eif2ak3 | 6336 | 0.126 | 0.5073 | No |
| 61 | Atf6 | 6493 | 0.121 | 0.5037 | No |
| 62 | Slc30a5 | 6496 | 0.121 | 0.5079 | No |
| 63 | Eif4e | 6924 | 0.104 | 0.4901 | No |
| 64 | Exoc2 | 7274 | 0.089 | 0.4757 | No |
| 65 | Atp6v0d1 | 7311 | 0.088 | 0.4770 | No |
| 66 | Paip1 | 7372 | 0.085 | 0.4769 | No |
| 67 | Nop56 | 7447 | 0.081 | 0.4761 | No |
| 68 | Vegfa | 7594 | 0.074 | 0.4714 | No |
| 69 | Slc1a4 | 7706 | 0.069 | 0.4682 | No |
| 70 | Fus | 7818 | 0.065 | 0.4649 | No |
| 71 | Nfyb | 7862 | 0.063 | 0.4650 | No |
| 72 | Eif4a3 | 7965 | 0.059 | 0.4619 | No |
| 73 | Srprb | 7999 | 0.058 | 0.4623 | No |
| 74 | Atf3 | 8529 | 0.035 | 0.4370 | No |
| 75 | Mthfd2 | 8574 | 0.032 | 0.4359 | No |
| 76 | Gemin4 | 8597 | 0.031 | 0.4359 | No |
| 77 | Cks1b | 8660 | 0.028 | 0.4338 | No |
| 78 | Pdia5 | 8980 | 0.013 | 0.4182 | No |
| 79 | Tubb2a | 9070 | 0.010 | 0.4141 | No |
| 80 | Herpud1 | 9255 | 0.002 | 0.4049 | No |
| 81 | Rps14 | 9262 | 0.001 | 0.4047 | No |
| 82 | Exosc9 | 9461 | -0.006 | 0.3949 | No |
| 83 | Ddx10 | 9661 | -0.015 | 0.3855 | No |
| 84 | Sec31a | 9742 | -0.019 | 0.3821 | No |
| 85 | Cnot4 | 9923 | -0.025 | 0.3739 | No |
| 86 | Dkc1 | 9956 | -0.026 | 0.3732 | No |
| 87 | Exosc1 | 10032 | -0.030 | 0.3705 | No |
| 88 | Pop4 | 10557 | -0.052 | 0.3460 | No |
| 89 | Atf4 | 11066 | -0.072 | 0.3231 | No |
| 90 | Nabp1 | 11393 | -0.085 | 0.3097 | No |
| 91 | Ern1 | 11454 | -0.088 | 0.3098 | No |
| 92 | Stc2 | 11462 | -0.088 | 0.3125 | No |
| 93 | Rrp9 | 11626 | -0.095 | 0.3077 | No |
| 94 | Chac1 | 12128 | -0.118 | 0.2867 | No |
| 95 | Wfs1 | 12500 | -0.134 | 0.2727 | No |
| 96 | Banf1 | 12577 | -0.138 | 0.2738 | No |
| 97 | Imp3 | 12920 | -0.154 | 0.2620 | No |
| 98 | Exosc4 | 13793 | -0.193 | 0.2250 | No |
| 99 | Nfya | 14073 | -0.206 | 0.2182 | No |
| 100 | Eif4ebp1 | 14212 | -0.213 | 0.2188 | No |
| 101 | Lsm4 | 14214 | -0.213 | 0.2262 | No |
| 102 | Spcs1 | 14747 | -0.240 | 0.2080 | No |
| 103 | Yif1a | 15233 | -0.264 | 0.1929 | No |
| 104 | H2ax | 15768 | -0.293 | 0.1764 | No |
| 105 | Exosc5 | 16071 | -0.311 | 0.1721 | No |
| 106 | Wipi1 | 16436 | -0.330 | 0.1654 | No |
| 107 | Kdelr3 | 17735 | -0.419 | 0.1150 | No |