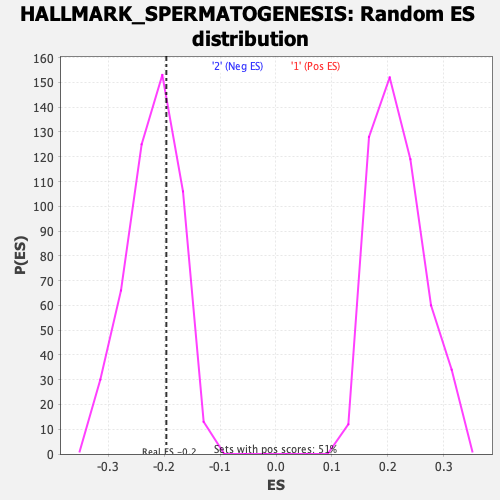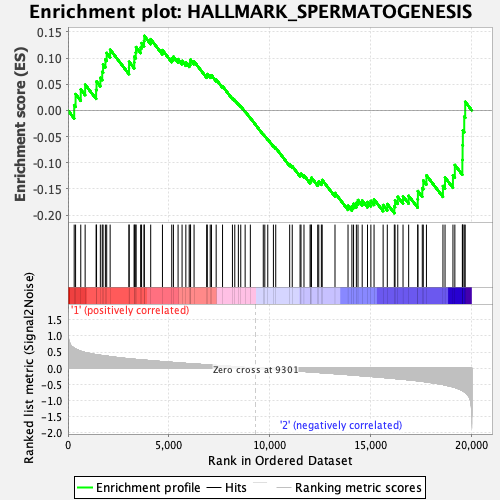
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | GMP.GMP.neu_Pheno.cls #Group2_versus_Group3.GMP.neu_Pheno.cls #Group2_versus_Group3_repos |
| Phenotype | GMP.neu_Pheno.cls#Group2_versus_Group3_repos |
| Upregulated in class | 2 |
| GeneSet | HALLMARK_SPERMATOGENESIS |
| Enrichment Score (ES) | -0.19634463 |
| Normalized Enrichment Score (NES) | -0.8937085 |
| Nominal p-value | 0.6680162 |
| FDR q-value | 0.79956317 |
| FWER p-Value | 0.997 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Nek2 | 307 | 0.616 | 0.0101 | No |
| 2 | Gmcl1 | 376 | 0.592 | 0.0311 | No |
| 3 | Art3 | 633 | 0.524 | 0.0399 | No |
| 4 | Cftr | 850 | 0.487 | 0.0492 | No |
| 5 | Bub1 | 1394 | 0.416 | 0.0392 | No |
| 6 | Stam2 | 1415 | 0.414 | 0.0553 | No |
| 7 | Pgs1 | 1604 | 0.398 | 0.0623 | No |
| 8 | Pacrg | 1697 | 0.391 | 0.0738 | No |
| 9 | Agfg1 | 1739 | 0.386 | 0.0877 | No |
| 10 | Ttk | 1854 | 0.377 | 0.0976 | No |
| 11 | Sirt1 | 1919 | 0.372 | 0.1097 | No |
| 12 | Jam3 | 2090 | 0.359 | 0.1160 | No |
| 13 | Pebp1 | 3026 | 0.290 | 0.0811 | No |
| 14 | Ncaph | 3029 | 0.290 | 0.0930 | No |
| 15 | Parp2 | 3284 | 0.275 | 0.0916 | No |
| 16 | Mtor | 3289 | 0.274 | 0.1028 | No |
| 17 | Hspa4l | 3360 | 0.271 | 0.1104 | No |
| 18 | Ide | 3374 | 0.270 | 0.1209 | No |
| 19 | Scg5 | 3603 | 0.258 | 0.1202 | No |
| 20 | Aurka | 3648 | 0.256 | 0.1285 | No |
| 21 | Dbf4 | 3772 | 0.248 | 0.1326 | No |
| 22 | Zc3h14 | 3783 | 0.248 | 0.1423 | No |
| 23 | Hspa1l | 4101 | 0.232 | 0.1360 | No |
| 24 | Nefh | 4684 | 0.201 | 0.1151 | No |
| 25 | Psmg1 | 5145 | 0.180 | 0.0995 | No |
| 26 | Cdk1 | 5228 | 0.176 | 0.1027 | No |
| 27 | Gfi1 | 5459 | 0.165 | 0.0979 | No |
| 28 | Ybx2 | 5657 | 0.156 | 0.0945 | No |
| 29 | Tle4 | 5844 | 0.147 | 0.0913 | No |
| 30 | Topbp1 | 6010 | 0.140 | 0.0888 | No |
| 31 | Kif2c | 6060 | 0.137 | 0.0920 | No |
| 32 | Clvs1 | 6078 | 0.137 | 0.0968 | No |
| 33 | Vdac3 | 6252 | 0.130 | 0.0935 | No |
| 34 | Dmc1 | 6877 | 0.106 | 0.0666 | No |
| 35 | Csnk2a2 | 6908 | 0.105 | 0.0694 | No |
| 36 | Dcc | 7053 | 0.098 | 0.0662 | No |
| 37 | Cnih2 | 7115 | 0.095 | 0.0671 | No |
| 38 | Chfr | 7348 | 0.086 | 0.0590 | No |
| 39 | Nf2 | 7664 | 0.071 | 0.0461 | No |
| 40 | Arl4a | 8155 | 0.051 | 0.0236 | No |
| 41 | Ldhc | 8271 | 0.045 | 0.0197 | No |
| 42 | Rfc4 | 8449 | 0.038 | 0.0124 | No |
| 43 | Tsn | 8567 | 0.033 | 0.0079 | No |
| 44 | Cct6b | 8785 | 0.022 | -0.0021 | No |
| 45 | Mllt10 | 9043 | 0.011 | -0.0145 | No |
| 46 | Ddx4 | 9684 | -0.016 | -0.0460 | No |
| 47 | Tulp2 | 9760 | -0.019 | -0.0489 | No |
| 48 | Slc2a5 | 9907 | -0.024 | -0.0553 | No |
| 49 | Strbp | 10187 | -0.037 | -0.0678 | No |
| 50 | Pias2 | 10302 | -0.042 | -0.0717 | No |
| 51 | Clpb | 10993 | -0.070 | -0.1035 | No |
| 52 | Ccnb2 | 11121 | -0.074 | -0.1068 | No |
| 53 | Ip6k1 | 11509 | -0.090 | -0.1225 | No |
| 54 | Rad17 | 11555 | -0.092 | -0.1209 | No |
| 55 | Slc12a2 | 11703 | -0.099 | -0.1242 | No |
| 56 | Nos1 | 12006 | -0.112 | -0.1347 | No |
| 57 | Ezh2 | 12048 | -0.114 | -0.1321 | No |
| 58 | Coil | 12074 | -0.116 | -0.1286 | No |
| 59 | Phf7 | 12384 | -0.128 | -0.1388 | No |
| 60 | Hspa2 | 12436 | -0.130 | -0.1359 | No |
| 61 | Spata6 | 12565 | -0.138 | -0.1367 | No |
| 62 | Mast2 | 12606 | -0.139 | -0.1329 | No |
| 63 | Pomc | 13241 | -0.167 | -0.1578 | No |
| 64 | Nphp1 | 13887 | -0.196 | -0.1821 | No |
| 65 | Acrbp | 14079 | -0.206 | -0.1832 | No |
| 66 | Adam2 | 14161 | -0.210 | -0.1785 | No |
| 67 | Braf | 14301 | -0.218 | -0.1765 | No |
| 68 | Mlf1 | 14382 | -0.223 | -0.1713 | No |
| 69 | Dmrt1 | 14590 | -0.231 | -0.1721 | No |
| 70 | Prkar2a | 14853 | -0.246 | -0.1751 | No |
| 71 | Taldo1 | 15020 | -0.253 | -0.1730 | No |
| 72 | Scg3 | 15178 | -0.260 | -0.1701 | No |
| 73 | Oaz3 | 15628 | -0.285 | -0.1808 | No |
| 74 | Camk4 | 15835 | -0.296 | -0.1789 | No |
| 75 | Tekt2 | 16183 | -0.316 | -0.1833 | Yes |
| 76 | Crisp2 | 16219 | -0.319 | -0.1719 | Yes |
| 77 | Zpbp | 16354 | -0.326 | -0.1651 | Yes |
| 78 | Ift88 | 16615 | -0.340 | -0.1642 | Yes |
| 79 | Gstm5 | 16894 | -0.357 | -0.1633 | Yes |
| 80 | Map7 | 17343 | -0.387 | -0.1698 | Yes |
| 81 | Septin4 | 17353 | -0.389 | -0.1542 | Yes |
| 82 | Phkg2 | 17570 | -0.404 | -0.1483 | Yes |
| 83 | Tnni3 | 17619 | -0.407 | -0.1339 | Yes |
| 84 | Grm8 | 17776 | -0.423 | -0.1243 | Yes |
| 85 | Cdkn3 | 18595 | -0.503 | -0.1445 | Yes |
| 86 | She | 18698 | -0.516 | -0.1283 | Yes |
| 87 | Zc2hc1c | 19091 | -0.576 | -0.1242 | Yes |
| 88 | Gapdhs | 19185 | -0.593 | -0.1043 | Yes |
| 89 | Adad1 | 19557 | -0.686 | -0.0946 | Yes |
| 90 | Tcp11 | 19569 | -0.690 | -0.0666 | Yes |
| 91 | Lpin1 | 19582 | -0.695 | -0.0385 | Yes |
| 92 | Il12rb2 | 19650 | -0.724 | -0.0119 | Yes |
| 93 | Pcsk4 | 19698 | -0.743 | 0.0165 | Yes |