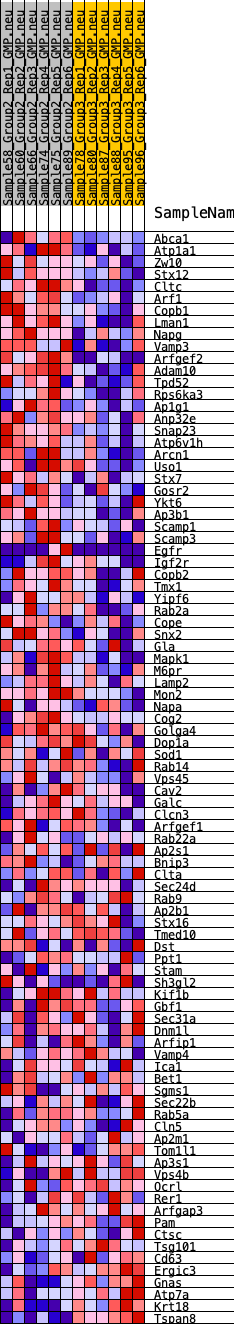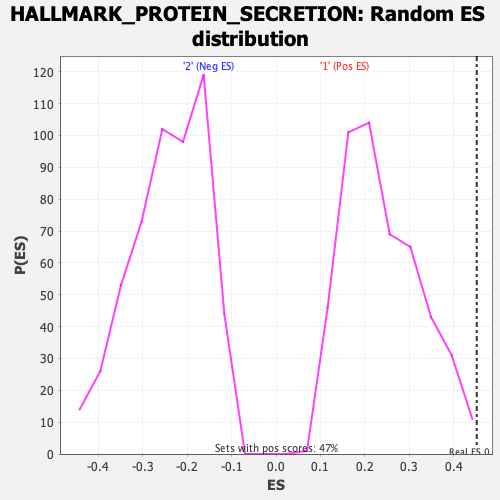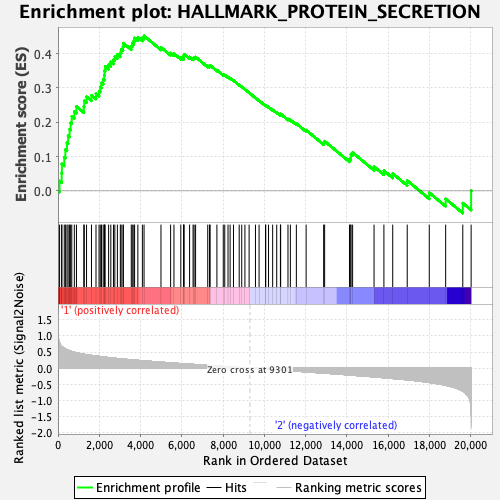
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | GMP.GMP.neu_Pheno.cls #Group2_versus_Group3.GMP.neu_Pheno.cls #Group2_versus_Group3_repos |
| Phenotype | GMP.neu_Pheno.cls#Group2_versus_Group3_repos |
| Upregulated in class | 1 |
| GeneSet | HALLMARK_PROTEIN_SECRETION |
| Enrichment Score (ES) | 0.45140722 |
| Normalized Enrichment Score (NES) | 1.8832803 |
| Nominal p-value | 0.006369427 |
| FDR q-value | 0.037520777 |
| FWER p-Value | 0.028 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Abca1 | 76 | 0.792 | 0.0287 | Yes |
| 2 | Atp1a1 | 176 | 0.676 | 0.0516 | Yes |
| 3 | Zw10 | 188 | 0.672 | 0.0786 | Yes |
| 4 | Stx12 | 303 | 0.618 | 0.0983 | Yes |
| 5 | Cltc | 358 | 0.598 | 0.1202 | Yes |
| 6 | Arf1 | 434 | 0.577 | 0.1401 | Yes |
| 7 | Copb1 | 490 | 0.563 | 0.1605 | Yes |
| 8 | Lman1 | 561 | 0.543 | 0.1794 | Yes |
| 9 | Napg | 612 | 0.529 | 0.1986 | Yes |
| 10 | Vamp3 | 667 | 0.516 | 0.2171 | Yes |
| 11 | Arfgef2 | 791 | 0.493 | 0.2312 | Yes |
| 12 | Adam10 | 894 | 0.480 | 0.2458 | Yes |
| 13 | Tpd52 | 1251 | 0.434 | 0.2458 | Yes |
| 14 | Rps6ka3 | 1272 | 0.430 | 0.2625 | Yes |
| 15 | Ap1g1 | 1383 | 0.417 | 0.2741 | Yes |
| 16 | Anp32e | 1622 | 0.396 | 0.2785 | Yes |
| 17 | Snap23 | 1841 | 0.378 | 0.2831 | Yes |
| 18 | Atp6v1h | 1989 | 0.367 | 0.2908 | Yes |
| 19 | Arcn1 | 2047 | 0.362 | 0.3028 | Yes |
| 20 | Uso1 | 2106 | 0.357 | 0.3145 | Yes |
| 21 | Stx7 | 2188 | 0.350 | 0.3249 | Yes |
| 22 | Gosr2 | 2242 | 0.346 | 0.3364 | Yes |
| 23 | Ykt6 | 2253 | 0.345 | 0.3501 | Yes |
| 24 | Ap3b1 | 2284 | 0.342 | 0.3626 | Yes |
| 25 | Scamp1 | 2452 | 0.328 | 0.3677 | Yes |
| 26 | Scamp3 | 2559 | 0.321 | 0.3756 | Yes |
| 27 | Egfr | 2687 | 0.313 | 0.3821 | Yes |
| 28 | Igf2r | 2753 | 0.308 | 0.3915 | Yes |
| 29 | Copb2 | 2877 | 0.299 | 0.3976 | Yes |
| 30 | Tmx1 | 3020 | 0.291 | 0.4024 | Yes |
| 31 | Yipf6 | 3059 | 0.289 | 0.4124 | Yes |
| 32 | Rab2a | 3149 | 0.282 | 0.4195 | Yes |
| 33 | Cope | 3168 | 0.281 | 0.4301 | Yes |
| 34 | Snx2 | 3550 | 0.261 | 0.4217 | Yes |
| 35 | Gla | 3599 | 0.258 | 0.4300 | Yes |
| 36 | Mapk1 | 3673 | 0.255 | 0.4368 | Yes |
| 37 | M6pr | 3711 | 0.253 | 0.4453 | Yes |
| 38 | Lamp2 | 3869 | 0.244 | 0.4474 | Yes |
| 39 | Mon2 | 4088 | 0.233 | 0.4461 | Yes |
| 40 | Napa | 4169 | 0.228 | 0.4514 | Yes |
| 41 | Cog2 | 4989 | 0.187 | 0.4180 | No |
| 42 | Golga4 | 5456 | 0.165 | 0.4014 | No |
| 43 | Dop1a | 5618 | 0.158 | 0.3999 | No |
| 44 | Sod1 | 5950 | 0.143 | 0.3891 | No |
| 45 | Rab14 | 6084 | 0.137 | 0.3881 | No |
| 46 | Vps45 | 6088 | 0.137 | 0.3935 | No |
| 47 | Cav2 | 6109 | 0.136 | 0.3981 | No |
| 48 | Galc | 6375 | 0.124 | 0.3899 | No |
| 49 | Clcn3 | 6544 | 0.119 | 0.3864 | No |
| 50 | Arfgef1 | 6593 | 0.116 | 0.3888 | No |
| 51 | Rab22a | 6666 | 0.114 | 0.3898 | No |
| 52 | Ap2s1 | 7255 | 0.090 | 0.3640 | No |
| 53 | Bnip3 | 7341 | 0.086 | 0.3633 | No |
| 54 | Clta | 7370 | 0.085 | 0.3654 | No |
| 55 | Sec24d | 7699 | 0.070 | 0.3518 | No |
| 56 | Rab9 | 8007 | 0.057 | 0.3388 | No |
| 57 | Ap2b1 | 8079 | 0.054 | 0.3374 | No |
| 58 | Stx16 | 8237 | 0.047 | 0.3315 | No |
| 59 | Tmed10 | 8339 | 0.042 | 0.3281 | No |
| 60 | Dst | 8502 | 0.036 | 0.3215 | No |
| 61 | Ppt1 | 8779 | 0.023 | 0.3086 | No |
| 62 | Stam | 8902 | 0.017 | 0.3032 | No |
| 63 | Sh3gl2 | 9056 | 0.010 | 0.2959 | No |
| 64 | Kif1b | 9259 | 0.001 | 0.2858 | No |
| 65 | Gbf1 | 9567 | -0.011 | 0.2709 | No |
| 66 | Sec31a | 9742 | -0.019 | 0.2629 | No |
| 67 | Dnm1l | 10053 | -0.031 | 0.2486 | No |
| 68 | Arfip1 | 10066 | -0.031 | 0.2493 | No |
| 69 | Vamp4 | 10200 | -0.038 | 0.2442 | No |
| 70 | Ica1 | 10400 | -0.046 | 0.2361 | No |
| 71 | Bet1 | 10597 | -0.054 | 0.2285 | No |
| 72 | Sgms1 | 10776 | -0.061 | 0.2221 | No |
| 73 | Sec22b | 10788 | -0.062 | 0.2240 | No |
| 74 | Rab5a | 11142 | -0.075 | 0.2094 | No |
| 75 | Cln5 | 11258 | -0.079 | 0.2069 | No |
| 76 | Ap2m1 | 11548 | -0.091 | 0.1962 | No |
| 77 | Tom1l1 | 12022 | -0.113 | 0.1771 | No |
| 78 | Ap3s1 | 12871 | -0.151 | 0.1408 | No |
| 79 | Vps4b | 12918 | -0.154 | 0.1448 | No |
| 80 | Ocrl | 14122 | -0.208 | 0.0930 | No |
| 81 | Rer1 | 14189 | -0.212 | 0.0984 | No |
| 82 | Arfgap3 | 14190 | -0.212 | 0.1071 | No |
| 83 | Pam | 14274 | -0.217 | 0.1118 | No |
| 84 | Ctsc | 15313 | -0.267 | 0.0707 | No |
| 85 | Tsg101 | 15790 | -0.294 | 0.0589 | No |
| 86 | Cd63 | 16217 | -0.318 | 0.0506 | No |
| 87 | Ergic3 | 16920 | -0.359 | 0.0302 | No |
| 88 | Gnas | 17988 | -0.442 | -0.0052 | No |
| 89 | Atp7a | 18782 | -0.529 | -0.0232 | No |
| 90 | Krt18 | 19612 | -0.710 | -0.0356 | No |
| 91 | Tspan8 | 20016 | -1.371 | 0.0005 | No |