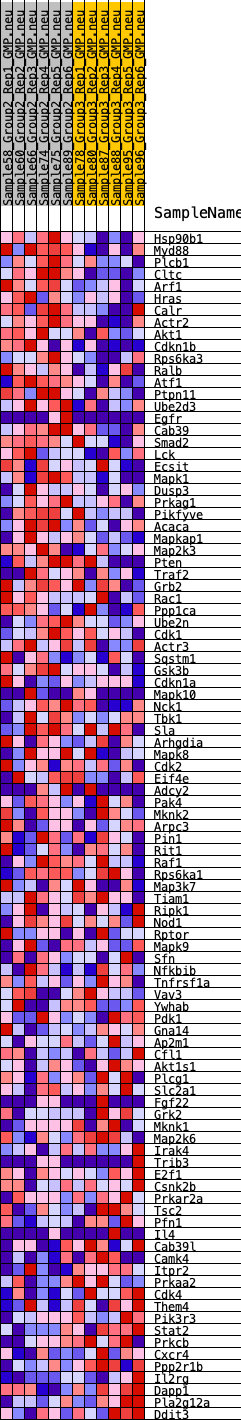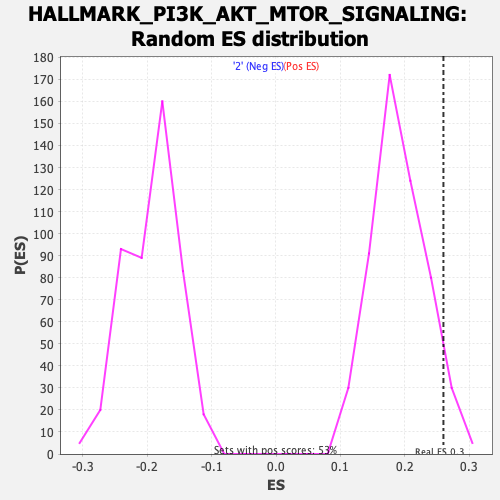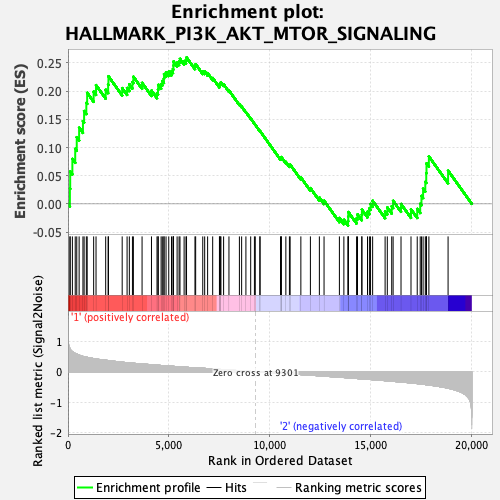
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | GMP.GMP.neu_Pheno.cls #Group2_versus_Group3.GMP.neu_Pheno.cls #Group2_versus_Group3_repos |
| Phenotype | GMP.neu_Pheno.cls#Group2_versus_Group3_repos |
| Upregulated in class | 1 |
| GeneSet | HALLMARK_PI3K_AKT_MTOR_SIGNALING |
| Enrichment Score (ES) | 0.25988343 |
| Normalized Enrichment Score (NES) | 1.3554959 |
| Nominal p-value | 0.060150377 |
| FDR q-value | 0.27841586 |
| FWER p-Value | 0.67 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Hsp90b1 | 94 | 0.763 | 0.0273 | Yes |
| 2 | Myd88 | 109 | 0.740 | 0.0576 | Yes |
| 3 | Plcb1 | 217 | 0.657 | 0.0798 | Yes |
| 4 | Cltc | 358 | 0.598 | 0.0979 | Yes |
| 5 | Arf1 | 434 | 0.577 | 0.1183 | Yes |
| 6 | Hras | 552 | 0.546 | 0.1353 | Yes |
| 7 | Calr | 741 | 0.503 | 0.1470 | Yes |
| 8 | Actr2 | 799 | 0.492 | 0.1648 | Yes |
| 9 | Akt1 | 909 | 0.478 | 0.1794 | Yes |
| 10 | Cdkn1b | 951 | 0.472 | 0.1971 | Yes |
| 11 | Rps6ka3 | 1272 | 0.430 | 0.1991 | Yes |
| 12 | Ralb | 1390 | 0.416 | 0.2107 | Yes |
| 13 | Atf1 | 1869 | 0.376 | 0.2025 | Yes |
| 14 | Ptpn11 | 1990 | 0.367 | 0.2118 | Yes |
| 15 | Ube2d3 | 2006 | 0.366 | 0.2264 | Yes |
| 16 | Egfr | 2687 | 0.313 | 0.2054 | Yes |
| 17 | Cab39 | 2930 | 0.296 | 0.2057 | Yes |
| 18 | Smad2 | 3042 | 0.290 | 0.2122 | Yes |
| 19 | Lck | 3196 | 0.280 | 0.2163 | Yes |
| 20 | Ecsit | 3242 | 0.278 | 0.2257 | Yes |
| 21 | Mapk1 | 3673 | 0.255 | 0.2148 | Yes |
| 22 | Dusp3 | 4140 | 0.230 | 0.2011 | Yes |
| 23 | Prkag1 | 4414 | 0.215 | 0.1964 | Yes |
| 24 | Pikfyve | 4464 | 0.212 | 0.2028 | Yes |
| 25 | Acaca | 4475 | 0.212 | 0.2112 | Yes |
| 26 | Mapkap1 | 4617 | 0.205 | 0.2127 | Yes |
| 27 | Map2k3 | 4677 | 0.201 | 0.2182 | Yes |
| 28 | Pten | 4756 | 0.196 | 0.2225 | Yes |
| 29 | Traf2 | 4765 | 0.195 | 0.2303 | Yes |
| 30 | Grb2 | 4862 | 0.191 | 0.2335 | Yes |
| 31 | Rac1 | 4996 | 0.187 | 0.2347 | Yes |
| 32 | Ppp1ca | 5133 | 0.181 | 0.2354 | Yes |
| 33 | Ube2n | 5205 | 0.177 | 0.2393 | Yes |
| 34 | Cdk1 | 5228 | 0.176 | 0.2456 | Yes |
| 35 | Actr3 | 5233 | 0.176 | 0.2527 | Yes |
| 36 | Sqstm1 | 5411 | 0.168 | 0.2509 | Yes |
| 37 | Gsk3b | 5508 | 0.163 | 0.2529 | Yes |
| 38 | Cdkn1a | 5553 | 0.161 | 0.2574 | Yes |
| 39 | Mapk10 | 5758 | 0.151 | 0.2535 | Yes |
| 40 | Nck1 | 5852 | 0.147 | 0.2550 | Yes |
| 41 | Tbk1 | 5878 | 0.146 | 0.2599 | Yes |
| 42 | Sla | 6297 | 0.128 | 0.2443 | No |
| 43 | Arhgdia | 6331 | 0.126 | 0.2479 | No |
| 44 | Mapk8 | 6683 | 0.113 | 0.2351 | No |
| 45 | Cdk2 | 6778 | 0.110 | 0.2349 | No |
| 46 | Eif4e | 6924 | 0.104 | 0.2320 | No |
| 47 | Adcy2 | 7175 | 0.092 | 0.2233 | No |
| 48 | Pak4 | 7510 | 0.078 | 0.2099 | No |
| 49 | Mknk2 | 7526 | 0.077 | 0.2123 | No |
| 50 | Arpc3 | 7554 | 0.076 | 0.2142 | No |
| 51 | Pin1 | 7584 | 0.075 | 0.2158 | No |
| 52 | Rit1 | 7717 | 0.069 | 0.2121 | No |
| 53 | Raf1 | 7982 | 0.058 | 0.2013 | No |
| 54 | Rps6ka1 | 8500 | 0.036 | 0.1769 | No |
| 55 | Map3k7 | 8611 | 0.030 | 0.1726 | No |
| 56 | Tiam1 | 8823 | 0.020 | 0.1629 | No |
| 57 | Ripk1 | 9060 | 0.010 | 0.1515 | No |
| 58 | Nod1 | 9245 | 0.002 | 0.1423 | No |
| 59 | Rptor | 9287 | 0.000 | 0.1403 | No |
| 60 | Mapk9 | 9516 | -0.009 | 0.1292 | No |
| 61 | Sfn | 9523 | -0.009 | 0.1292 | No |
| 62 | Nfkbib | 10539 | -0.052 | 0.0805 | No |
| 63 | Tnfrsf1a | 10576 | -0.053 | 0.0809 | No |
| 64 | Vav3 | 10577 | -0.053 | 0.0831 | No |
| 65 | Ywhab | 10804 | -0.062 | 0.0744 | No |
| 66 | Pdk1 | 10981 | -0.070 | 0.0685 | No |
| 67 | Gna14 | 11016 | -0.071 | 0.0697 | No |
| 68 | Ap2m1 | 11548 | -0.091 | 0.0469 | No |
| 69 | Cfl1 | 12021 | -0.113 | 0.0280 | No |
| 70 | Akt1s1 | 12466 | -0.132 | 0.0112 | No |
| 71 | Plcg1 | 12698 | -0.144 | 0.0057 | No |
| 72 | Slc2a1 | 13459 | -0.178 | -0.0250 | No |
| 73 | Fgf22 | 13681 | -0.188 | -0.0282 | No |
| 74 | Grk2 | 13888 | -0.196 | -0.0303 | No |
| 75 | Mknk1 | 13898 | -0.197 | -0.0225 | No |
| 76 | Map2k6 | 13903 | -0.197 | -0.0144 | No |
| 77 | Irak4 | 14311 | -0.219 | -0.0257 | No |
| 78 | Trib3 | 14357 | -0.221 | -0.0186 | No |
| 79 | E2f1 | 14569 | -0.230 | -0.0196 | No |
| 80 | Csnk2b | 14576 | -0.231 | -0.0102 | No |
| 81 | Prkar2a | 14853 | -0.246 | -0.0137 | No |
| 82 | Tsc2 | 14947 | -0.250 | -0.0079 | No |
| 83 | Pfn1 | 15004 | -0.252 | -0.0001 | No |
| 84 | Il4 | 15108 | -0.258 | 0.0055 | No |
| 85 | Cab39l | 15726 | -0.290 | -0.0133 | No |
| 86 | Camk4 | 15835 | -0.296 | -0.0063 | No |
| 87 | Itpr2 | 16054 | -0.310 | -0.0042 | No |
| 88 | Prkaa2 | 16123 | -0.314 | 0.0055 | No |
| 89 | Cdk4 | 16515 | -0.333 | -0.0001 | No |
| 90 | Them4 | 17010 | -0.365 | -0.0096 | No |
| 91 | Pik3r3 | 17319 | -0.386 | -0.0089 | No |
| 92 | Stat2 | 17467 | -0.397 | 0.0004 | No |
| 93 | Prkcb | 17529 | -0.401 | 0.0141 | No |
| 94 | Cxcr4 | 17610 | -0.407 | 0.0272 | No |
| 95 | Ppp2r1b | 17719 | -0.418 | 0.0393 | No |
| 96 | Il2rg | 17767 | -0.423 | 0.0547 | No |
| 97 | Dapp1 | 17779 | -0.424 | 0.0719 | No |
| 98 | Pla2g12a | 17897 | -0.434 | 0.0842 | No |
| 99 | Ddit3 | 18849 | -0.539 | 0.0591 | No |