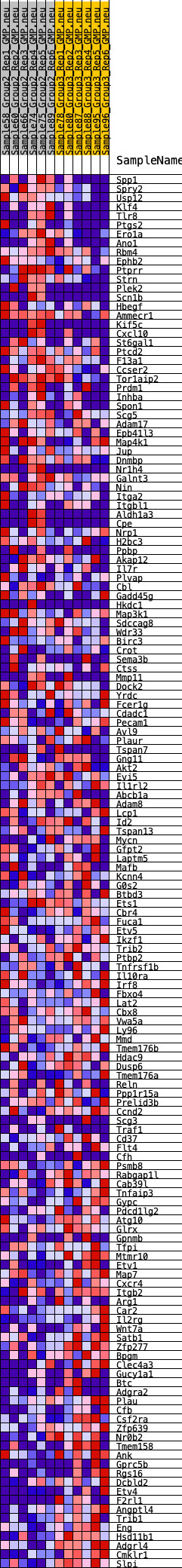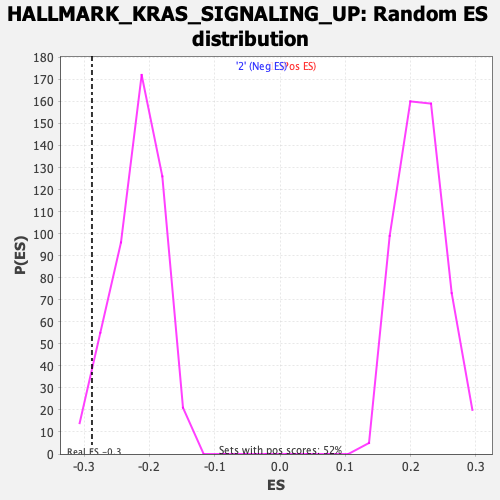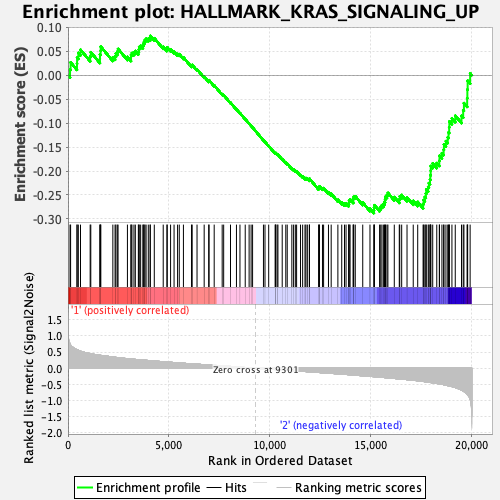
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | GMP.GMP.neu_Pheno.cls #Group2_versus_Group3.GMP.neu_Pheno.cls #Group2_versus_Group3_repos |
| Phenotype | GMP.neu_Pheno.cls#Group2_versus_Group3_repos |
| Upregulated in class | 2 |
| GeneSet | HALLMARK_KRAS_SIGNALING_UP |
| Enrichment Score (ES) | -0.2879306 |
| Normalized Enrichment Score (NES) | -1.3198956 |
| Nominal p-value | 0.02892562 |
| FDR q-value | 0.78432184 |
| FWER p-Value | 0.736 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Spp1 | 101 | 0.754 | 0.0121 | No |
| 2 | Spry2 | 129 | 0.713 | 0.0270 | No |
| 3 | Usp12 | 445 | 0.574 | 0.0242 | No |
| 4 | Klf4 | 451 | 0.573 | 0.0370 | No |
| 5 | Tlr8 | 512 | 0.557 | 0.0467 | No |
| 6 | Ptgs2 | 620 | 0.527 | 0.0533 | No |
| 7 | Ero1a | 1099 | 0.452 | 0.0395 | No |
| 8 | Ano1 | 1130 | 0.447 | 0.0482 | No |
| 9 | Rbm4 | 1584 | 0.400 | 0.0345 | No |
| 10 | Ephb2 | 1586 | 0.400 | 0.0436 | No |
| 11 | Ptprr | 1618 | 0.397 | 0.0511 | No |
| 12 | Strn | 1623 | 0.396 | 0.0599 | No |
| 13 | Plek2 | 2224 | 0.347 | 0.0376 | No |
| 14 | Scn1b | 2334 | 0.339 | 0.0398 | No |
| 15 | Hbegf | 2373 | 0.336 | 0.0456 | No |
| 16 | Ammecr1 | 2444 | 0.329 | 0.0495 | No |
| 17 | Kif5c | 2482 | 0.324 | 0.0551 | No |
| 18 | Cxcl10 | 2947 | 0.295 | 0.0384 | No |
| 19 | St6gal1 | 3125 | 0.284 | 0.0360 | No |
| 20 | Ptcd2 | 3126 | 0.284 | 0.0424 | No |
| 21 | F13a1 | 3171 | 0.281 | 0.0466 | No |
| 22 | Ccser2 | 3271 | 0.276 | 0.0479 | No |
| 23 | Tor1aip2 | 3339 | 0.272 | 0.0508 | No |
| 24 | Prdm1 | 3489 | 0.265 | 0.0493 | No |
| 25 | Inhba | 3515 | 0.263 | 0.0540 | No |
| 26 | Spon1 | 3530 | 0.262 | 0.0593 | No |
| 27 | Scg5 | 3603 | 0.258 | 0.0615 | No |
| 28 | Adam17 | 3715 | 0.252 | 0.0617 | No |
| 29 | Epb41l3 | 3736 | 0.251 | 0.0664 | No |
| 30 | Map4k1 | 3775 | 0.248 | 0.0702 | No |
| 31 | Jup | 3815 | 0.246 | 0.0738 | No |
| 32 | Dnmbp | 3872 | 0.243 | 0.0765 | No |
| 33 | Nr1h4 | 3982 | 0.239 | 0.0765 | No |
| 34 | Galnt3 | 4073 | 0.234 | 0.0773 | No |
| 35 | Nin | 4080 | 0.233 | 0.0823 | No |
| 36 | Itga2 | 4279 | 0.222 | 0.0774 | No |
| 37 | Itgbl1 | 4722 | 0.198 | 0.0596 | No |
| 38 | Aldh1a3 | 4907 | 0.189 | 0.0547 | No |
| 39 | Cpe | 4920 | 0.189 | 0.0584 | No |
| 40 | Nrp1 | 5087 | 0.183 | 0.0542 | No |
| 41 | H2bc3 | 5260 | 0.174 | 0.0495 | No |
| 42 | Ppbp | 5434 | 0.167 | 0.0446 | No |
| 43 | Akap12 | 5518 | 0.162 | 0.0441 | No |
| 44 | Il7r | 5724 | 0.153 | 0.0373 | No |
| 45 | Plvap | 6124 | 0.135 | 0.0203 | No |
| 46 | Cbl | 6158 | 0.133 | 0.0217 | No |
| 47 | Gadd45g | 6399 | 0.123 | 0.0124 | No |
| 48 | Hkdc1 | 6753 | 0.110 | -0.0028 | No |
| 49 | Map3k1 | 6979 | 0.101 | -0.0119 | No |
| 50 | Sdccag8 | 6995 | 0.100 | -0.0103 | No |
| 51 | Wdr33 | 7250 | 0.090 | -0.0211 | No |
| 52 | Birc3 | 7646 | 0.072 | -0.0393 | No |
| 53 | Crot | 7720 | 0.069 | -0.0414 | No |
| 54 | Sema3b | 8059 | 0.055 | -0.0572 | No |
| 55 | Ctss | 8354 | 0.042 | -0.0710 | No |
| 56 | Mmp11 | 8528 | 0.035 | -0.0789 | No |
| 57 | Dock2 | 8789 | 0.022 | -0.0915 | No |
| 58 | Yrdc | 8982 | 0.013 | -0.1009 | No |
| 59 | Fcer1g | 9081 | 0.009 | -0.1056 | No |
| 60 | Cdadc1 | 9154 | 0.006 | -0.1091 | No |
| 61 | Pecam1 | 9693 | -0.016 | -0.1358 | No |
| 62 | Avl9 | 9769 | -0.020 | -0.1391 | No |
| 63 | Plaur | 9953 | -0.026 | -0.1477 | No |
| 64 | Tspan7 | 10272 | -0.041 | -0.1628 | No |
| 65 | Gng11 | 10295 | -0.042 | -0.1629 | No |
| 66 | Akt2 | 10299 | -0.042 | -0.1621 | No |
| 67 | Evi5 | 10370 | -0.045 | -0.1646 | No |
| 68 | Il1rl2 | 10411 | -0.047 | -0.1656 | No |
| 69 | Abcb1a | 10623 | -0.055 | -0.1750 | No |
| 70 | Adam8 | 10801 | -0.062 | -0.1824 | No |
| 71 | Lcp1 | 10872 | -0.065 | -0.1845 | No |
| 72 | Id2 | 11101 | -0.073 | -0.1943 | No |
| 73 | Tspan13 | 11188 | -0.077 | -0.1969 | No |
| 74 | Mycn | 11274 | -0.080 | -0.1993 | No |
| 75 | Gfpt2 | 11337 | -0.083 | -0.2006 | No |
| 76 | Laptm5 | 11531 | -0.091 | -0.2082 | No |
| 77 | Mafb | 11637 | -0.096 | -0.2113 | No |
| 78 | Kcnn4 | 11753 | -0.100 | -0.2148 | No |
| 79 | G0s2 | 11798 | -0.103 | -0.2147 | No |
| 80 | Btbd3 | 11869 | -0.107 | -0.2158 | No |
| 81 | Ets1 | 11967 | -0.110 | -0.2182 | No |
| 82 | Cbr4 | 11976 | -0.111 | -0.2160 | No |
| 83 | Fuca1 | 12428 | -0.130 | -0.2358 | No |
| 84 | Etv5 | 12452 | -0.131 | -0.2339 | No |
| 85 | Ikzf1 | 12471 | -0.132 | -0.2318 | No |
| 86 | Trib2 | 12620 | -0.140 | -0.2361 | No |
| 87 | Ptbp2 | 12676 | -0.143 | -0.2356 | No |
| 88 | Tnfrsf1b | 12917 | -0.154 | -0.2442 | No |
| 89 | Il10ra | 13052 | -0.159 | -0.2473 | No |
| 90 | Irf8 | 13384 | -0.174 | -0.2600 | No |
| 91 | Fbxo4 | 13575 | -0.183 | -0.2654 | No |
| 92 | Lat2 | 13712 | -0.189 | -0.2679 | No |
| 93 | Cbx8 | 13789 | -0.193 | -0.2674 | No |
| 94 | Vwa5a | 13918 | -0.198 | -0.2693 | No |
| 95 | Ly96 | 13934 | -0.199 | -0.2655 | No |
| 96 | Mmd | 13939 | -0.199 | -0.2612 | No |
| 97 | Tmem176b | 13989 | -0.201 | -0.2591 | No |
| 98 | Hdac9 | 14146 | -0.209 | -0.2622 | No |
| 99 | Dusp6 | 14147 | -0.209 | -0.2574 | No |
| 100 | Tmem176a | 14171 | -0.211 | -0.2537 | No |
| 101 | Reln | 14246 | -0.215 | -0.2526 | No |
| 102 | Ppp1r15a | 14617 | -0.233 | -0.2659 | No |
| 103 | Prelid3b | 14976 | -0.251 | -0.2782 | No |
| 104 | Ccnd2 | 15171 | -0.260 | -0.2820 | Yes |
| 105 | Scg3 | 15178 | -0.260 | -0.2764 | Yes |
| 106 | Traf1 | 15197 | -0.261 | -0.2713 | Yes |
| 107 | Cd37 | 15452 | -0.275 | -0.2779 | Yes |
| 108 | Flt4 | 15510 | -0.278 | -0.2744 | Yes |
| 109 | Cfh | 15571 | -0.282 | -0.2710 | Yes |
| 110 | Psmb8 | 15653 | -0.286 | -0.2686 | Yes |
| 111 | Rabgap1l | 15697 | -0.289 | -0.2641 | Yes |
| 112 | Cab39l | 15726 | -0.290 | -0.2590 | Yes |
| 113 | Tnfaip3 | 15746 | -0.291 | -0.2533 | Yes |
| 114 | Gypc | 15817 | -0.295 | -0.2501 | Yes |
| 115 | Pdcd1lg2 | 15864 | -0.298 | -0.2456 | Yes |
| 116 | Atg10 | 16181 | -0.316 | -0.2543 | Yes |
| 117 | Glrx | 16429 | -0.329 | -0.2592 | Yes |
| 118 | Gpnmb | 16452 | -0.331 | -0.2528 | Yes |
| 119 | Tfpi | 16548 | -0.335 | -0.2499 | Yes |
| 120 | Mtmr10 | 16813 | -0.352 | -0.2552 | Yes |
| 121 | Etv1 | 17121 | -0.372 | -0.2622 | Yes |
| 122 | Map7 | 17343 | -0.387 | -0.2645 | Yes |
| 123 | Cxcr4 | 17610 | -0.407 | -0.2686 | Yes |
| 124 | Itgb2 | 17635 | -0.410 | -0.2605 | Yes |
| 125 | Arg1 | 17698 | -0.416 | -0.2541 | Yes |
| 126 | Car2 | 17746 | -0.420 | -0.2469 | Yes |
| 127 | Il2rg | 17767 | -0.423 | -0.2383 | Yes |
| 128 | Wnt7a | 17862 | -0.431 | -0.2332 | Yes |
| 129 | Satb1 | 17898 | -0.434 | -0.2251 | Yes |
| 130 | Zfp277 | 17953 | -0.439 | -0.2178 | Yes |
| 131 | Bpgm | 17971 | -0.440 | -0.2086 | Yes |
| 132 | Clec4a3 | 17987 | -0.442 | -0.1993 | Yes |
| 133 | Gucy1a1 | 17991 | -0.442 | -0.1894 | Yes |
| 134 | Btc | 18087 | -0.450 | -0.1839 | Yes |
| 135 | Adgra2 | 18288 | -0.470 | -0.1833 | Yes |
| 136 | Plau | 18414 | -0.484 | -0.1785 | Yes |
| 137 | Cfb | 18424 | -0.485 | -0.1679 | Yes |
| 138 | Csf2ra | 18544 | -0.497 | -0.1626 | Yes |
| 139 | Zfp639 | 18630 | -0.507 | -0.1553 | Yes |
| 140 | Nr0b2 | 18644 | -0.509 | -0.1444 | Yes |
| 141 | Tmem158 | 18743 | -0.522 | -0.1374 | Yes |
| 142 | Ank | 18833 | -0.537 | -0.1297 | Yes |
| 143 | Gprc5b | 18872 | -0.541 | -0.1192 | Yes |
| 144 | Rgs16 | 18904 | -0.545 | -0.1084 | Yes |
| 145 | Dcbld2 | 18913 | -0.547 | -0.0964 | Yes |
| 146 | Etv4 | 19040 | -0.567 | -0.0898 | Yes |
| 147 | F2rl1 | 19205 | -0.598 | -0.0844 | Yes |
| 148 | Angptl4 | 19515 | -0.673 | -0.0846 | Yes |
| 149 | Trib1 | 19598 | -0.702 | -0.0728 | Yes |
| 150 | Eng | 19635 | -0.722 | -0.0581 | Yes |
| 151 | Hsd11b1 | 19796 | -0.809 | -0.0478 | Yes |
| 152 | Adgrl4 | 19806 | -0.813 | -0.0297 | Yes |
| 153 | Cmklr1 | 19820 | -0.827 | -0.0115 | Yes |
| 154 | Slpi | 19943 | -0.957 | 0.0042 | Yes |