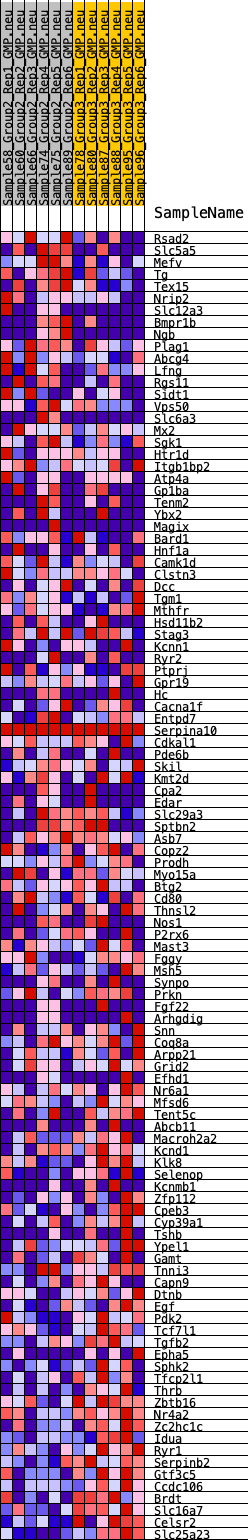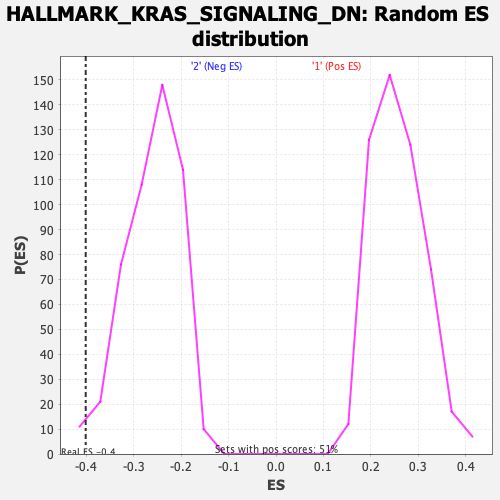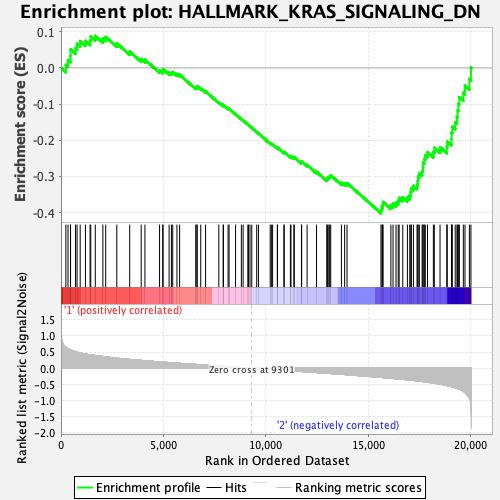
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | GMP.GMP.neu_Pheno.cls #Group2_versus_Group3.GMP.neu_Pheno.cls #Group2_versus_Group3_repos |
| Phenotype | GMP.neu_Pheno.cls#Group2_versus_Group3_repos |
| Upregulated in class | 2 |
| GeneSet | HALLMARK_KRAS_SIGNALING_DN |
| Enrichment Score (ES) | -0.40169603 |
| Normalized Enrichment Score (NES) | -1.542888 |
| Nominal p-value | 0.016393442 |
| FDR q-value | 0.60274845 |
| FWER p-Value | 0.347 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Rsad2 | 237 | 0.648 | 0.0083 | No |
| 2 | Slc5a5 | 342 | 0.604 | 0.0219 | No |
| 3 | Mefv | 464 | 0.569 | 0.0336 | No |
| 4 | Tg | 466 | 0.568 | 0.0513 | No |
| 5 | Tex15 | 709 | 0.509 | 0.0550 | No |
| 6 | Nrip2 | 783 | 0.495 | 0.0668 | No |
| 7 | Slc12a3 | 936 | 0.474 | 0.0739 | No |
| 8 | Bmpr1b | 1192 | 0.441 | 0.0749 | No |
| 9 | Ngb | 1418 | 0.414 | 0.0765 | No |
| 10 | Plag1 | 1452 | 0.411 | 0.0877 | No |
| 11 | Abcg4 | 1673 | 0.393 | 0.0889 | No |
| 12 | Lfng | 2043 | 0.362 | 0.0817 | No |
| 13 | Rgs11 | 2181 | 0.351 | 0.0857 | No |
| 14 | Sidt1 | 2722 | 0.311 | 0.0683 | No |
| 15 | Vps50 | 3352 | 0.271 | 0.0452 | No |
| 16 | Slc6a3 | 3912 | 0.241 | 0.0246 | No |
| 17 | Mx2 | 4095 | 0.233 | 0.0228 | No |
| 18 | Sgk1 | 4808 | 0.194 | -0.0069 | No |
| 19 | Htr1d | 4965 | 0.189 | -0.0089 | No |
| 20 | Itgb1bp2 | 4979 | 0.188 | -0.0037 | No |
| 21 | Atp4a | 5274 | 0.174 | -0.0130 | No |
| 22 | Gp1ba | 5387 | 0.169 | -0.0134 | No |
| 23 | Tenm2 | 5451 | 0.165 | -0.0114 | No |
| 24 | Ybx2 | 5657 | 0.156 | -0.0168 | No |
| 25 | Magix | 5787 | 0.150 | -0.0186 | No |
| 26 | Bard1 | 6573 | 0.117 | -0.0544 | No |
| 27 | Hnf1a | 6631 | 0.115 | -0.0536 | No |
| 28 | Camk1d | 6643 | 0.115 | -0.0506 | No |
| 29 | Clstn3 | 6817 | 0.108 | -0.0559 | No |
| 30 | Dcc | 7053 | 0.098 | -0.0647 | No |
| 31 | Tgm1 | 7700 | 0.070 | -0.0949 | No |
| 32 | Mthfr | 7912 | 0.061 | -0.1036 | No |
| 33 | Hsd11b2 | 7924 | 0.060 | -0.1023 | No |
| 34 | Stag3 | 8152 | 0.051 | -0.1121 | No |
| 35 | Kcnn1 | 8199 | 0.049 | -0.1129 | No |
| 36 | Ryr2 | 8517 | 0.035 | -0.1277 | No |
| 37 | Ptprj | 8799 | 0.022 | -0.1412 | No |
| 38 | Gpr19 | 8885 | 0.018 | -0.1449 | No |
| 39 | Hc | 9117 | 0.007 | -0.1562 | No |
| 40 | Cacna1f | 9144 | 0.006 | -0.1573 | No |
| 41 | Entpd7 | 9222 | 0.003 | -0.1611 | No |
| 42 | Serpina10 | 9308 | 0.000 | -0.1654 | No |
| 43 | Cdkal1 | 9548 | -0.010 | -0.1771 | No |
| 44 | Pde6b | 9633 | -0.014 | -0.1809 | No |
| 45 | Skil | 10207 | -0.038 | -0.2084 | No |
| 46 | Kmt2d | 10263 | -0.040 | -0.2100 | No |
| 47 | Cpa2 | 10324 | -0.043 | -0.2116 | No |
| 48 | Edar | 10555 | -0.052 | -0.2215 | No |
| 49 | Slc29a3 | 10561 | -0.052 | -0.2202 | No |
| 50 | Sptbn2 | 10875 | -0.065 | -0.2338 | No |
| 51 | Asb7 | 10890 | -0.066 | -0.2325 | No |
| 52 | Copz2 | 11191 | -0.077 | -0.2451 | No |
| 53 | Prodh | 11219 | -0.078 | -0.2441 | No |
| 54 | Myo15a | 11362 | -0.084 | -0.2486 | No |
| 55 | Btg2 | 11381 | -0.085 | -0.2468 | No |
| 56 | Cd80 | 11731 | -0.100 | -0.2613 | No |
| 57 | Thnsl2 | 11741 | -0.100 | -0.2586 | No |
| 58 | Nos1 | 12006 | -0.112 | -0.2683 | No |
| 59 | P2rx6 | 12467 | -0.132 | -0.2873 | No |
| 60 | Mast3 | 12966 | -0.155 | -0.3075 | No |
| 61 | Fggy | 12997 | -0.156 | -0.3041 | No |
| 62 | Msh5 | 13064 | -0.159 | -0.3025 | No |
| 63 | Synpo | 13109 | -0.161 | -0.2996 | No |
| 64 | Prkn | 13159 | -0.163 | -0.2970 | No |
| 65 | Fgf22 | 13681 | -0.188 | -0.3173 | No |
| 66 | Arhgdig | 13838 | -0.195 | -0.3191 | No |
| 67 | Snn | 13954 | -0.199 | -0.3186 | No |
| 68 | Coq8a | 15610 | -0.284 | -0.3928 | Yes |
| 69 | Arpp21 | 15660 | -0.286 | -0.3864 | Yes |
| 70 | Grid2 | 15691 | -0.288 | -0.3789 | Yes |
| 71 | Efhd1 | 15712 | -0.290 | -0.3709 | Yes |
| 72 | Nr6a1 | 16095 | -0.312 | -0.3803 | Yes |
| 73 | Mfsd6 | 16199 | -0.318 | -0.3756 | Yes |
| 74 | Tent5c | 16345 | -0.325 | -0.3727 | Yes |
| 75 | Abcb11 | 16451 | -0.331 | -0.3677 | Yes |
| 76 | Macroh2a2 | 16497 | -0.332 | -0.3596 | Yes |
| 77 | Kcnd1 | 16674 | -0.343 | -0.3577 | Yes |
| 78 | Klk8 | 16900 | -0.357 | -0.3578 | Yes |
| 79 | Selenop | 17013 | -0.365 | -0.3521 | Yes |
| 80 | Kcnmb1 | 17066 | -0.369 | -0.3432 | Yes |
| 81 | Zfp112 | 17096 | -0.370 | -0.3331 | Yes |
| 82 | Cpeb3 | 17194 | -0.377 | -0.3262 | Yes |
| 83 | Cyp39a1 | 17370 | -0.390 | -0.3228 | Yes |
| 84 | Tshb | 17408 | -0.392 | -0.3125 | Yes |
| 85 | Ypel1 | 17421 | -0.393 | -0.3008 | Yes |
| 86 | Gamt | 17482 | -0.398 | -0.2914 | Yes |
| 87 | Tnni3 | 17619 | -0.407 | -0.2856 | Yes |
| 88 | Capn9 | 17660 | -0.412 | -0.2747 | Yes |
| 89 | Dtnb | 17666 | -0.413 | -0.2621 | Yes |
| 90 | Egf | 17727 | -0.419 | -0.2520 | Yes |
| 91 | Pdk2 | 17773 | -0.423 | -0.2411 | Yes |
| 92 | Tcf7l1 | 17880 | -0.432 | -0.2330 | Yes |
| 93 | Tgfb2 | 18166 | -0.457 | -0.2330 | Yes |
| 94 | Epha5 | 18213 | -0.462 | -0.2209 | Yes |
| 95 | Sphk2 | 18492 | -0.492 | -0.2195 | Yes |
| 96 | Tfcp2l1 | 18821 | -0.535 | -0.2193 | Yes |
| 97 | Thrb | 18843 | -0.538 | -0.2035 | Yes |
| 98 | Zbtb16 | 19049 | -0.569 | -0.1961 | Yes |
| 99 | Nr4a2 | 19055 | -0.570 | -0.1785 | Yes |
| 100 | Zc2hc1c | 19091 | -0.576 | -0.1623 | Yes |
| 101 | Idua | 19232 | -0.602 | -0.1506 | Yes |
| 102 | Ryr1 | 19316 | -0.623 | -0.1353 | Yes |
| 103 | Serpinb2 | 19349 | -0.631 | -0.1172 | Yes |
| 104 | Gtf3c5 | 19386 | -0.640 | -0.0991 | Yes |
| 105 | Ccdc106 | 19435 | -0.650 | -0.0812 | Yes |
| 106 | Brdt | 19628 | -0.717 | -0.0685 | Yes |
| 107 | Slc16a7 | 19709 | -0.749 | -0.0491 | Yes |
| 108 | Celsr2 | 19924 | -0.925 | -0.0310 | Yes |
| 109 | Slc25a23 | 19999 | -1.158 | 0.0014 | Yes |