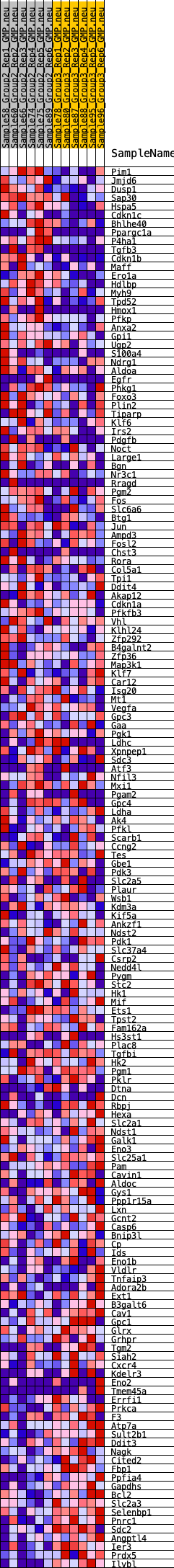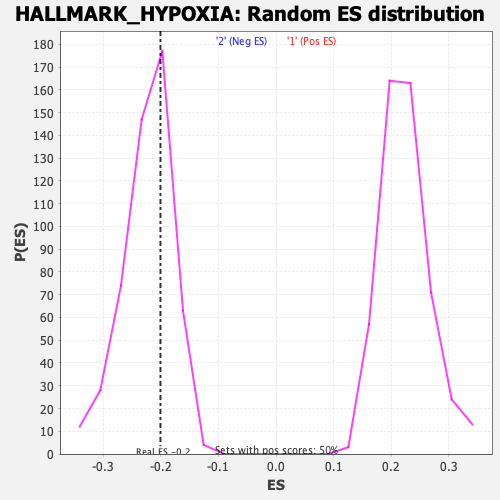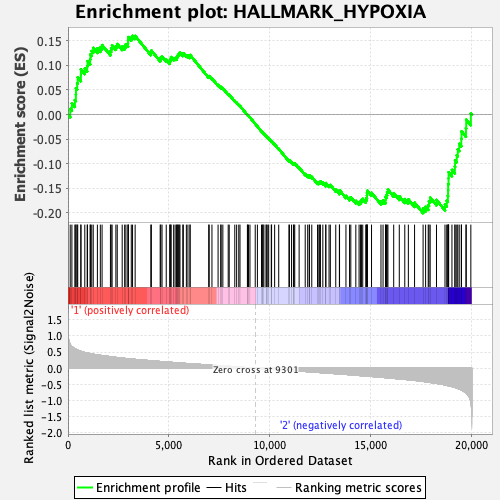
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | GMP.GMP.neu_Pheno.cls #Group2_versus_Group3.GMP.neu_Pheno.cls #Group2_versus_Group3_repos |
| Phenotype | GMP.neu_Pheno.cls#Group2_versus_Group3_repos |
| Upregulated in class | 2 |
| GeneSet | HALLMARK_HYPOXIA |
| Enrichment Score (ES) | -0.20074287 |
| Normalized Enrichment Score (NES) | -0.8962299 |
| Nominal p-value | 0.6970297 |
| FDR q-value | 0.83781976 |
| FWER p-Value | 0.997 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Pim1 | 110 | 0.739 | 0.0115 | No |
| 2 | Jmjd6 | 193 | 0.668 | 0.0228 | No |
| 3 | Dusp1 | 340 | 0.605 | 0.0294 | No |
| 4 | Sap30 | 390 | 0.586 | 0.0405 | No |
| 5 | Hspa5 | 395 | 0.586 | 0.0538 | No |
| 6 | Cdkn1c | 456 | 0.571 | 0.0639 | No |
| 7 | Bhlhe40 | 482 | 0.565 | 0.0757 | No |
| 8 | Ppargc1a | 639 | 0.523 | 0.0799 | No |
| 9 | P4ha1 | 640 | 0.523 | 0.0920 | No |
| 10 | Tgfb3 | 833 | 0.488 | 0.0936 | No |
| 11 | Cdkn1b | 951 | 0.472 | 0.0986 | No |
| 12 | Maff | 968 | 0.469 | 0.1086 | No |
| 13 | Ero1a | 1099 | 0.452 | 0.1125 | No |
| 14 | Hdlbp | 1110 | 0.450 | 0.1224 | No |
| 15 | Myh9 | 1177 | 0.443 | 0.1293 | No |
| 16 | Tpd52 | 1251 | 0.434 | 0.1356 | No |
| 17 | Hmox1 | 1457 | 0.411 | 0.1348 | No |
| 18 | Pfkp | 1603 | 0.399 | 0.1367 | No |
| 19 | Anxa2 | 1690 | 0.391 | 0.1414 | No |
| 20 | Gpi1 | 2104 | 0.357 | 0.1288 | No |
| 21 | Ugp2 | 2138 | 0.353 | 0.1353 | No |
| 22 | S100a4 | 2187 | 0.351 | 0.1410 | No |
| 23 | Ndrg1 | 2375 | 0.335 | 0.1393 | No |
| 24 | Aldoa | 2442 | 0.329 | 0.1436 | No |
| 25 | Egfr | 2687 | 0.313 | 0.1385 | No |
| 26 | Phkg1 | 2810 | 0.304 | 0.1394 | No |
| 27 | Foxo3 | 2872 | 0.299 | 0.1432 | No |
| 28 | Plin2 | 2978 | 0.293 | 0.1447 | No |
| 29 | Tiparp | 2980 | 0.293 | 0.1514 | No |
| 30 | Klf6 | 2992 | 0.292 | 0.1576 | No |
| 31 | Irs2 | 3138 | 0.282 | 0.1568 | No |
| 32 | Pdgfb | 3201 | 0.279 | 0.1601 | No |
| 33 | Noct | 3330 | 0.273 | 0.1600 | No |
| 34 | Large1 | 4108 | 0.231 | 0.1262 | No |
| 35 | Bgn | 4134 | 0.230 | 0.1303 | No |
| 36 | Nr3c1 | 4572 | 0.207 | 0.1131 | No |
| 37 | Rragd | 4600 | 0.206 | 0.1164 | No |
| 38 | Pgm2 | 4663 | 0.201 | 0.1180 | No |
| 39 | Fos | 4872 | 0.191 | 0.1119 | No |
| 40 | Slc6a6 | 5048 | 0.185 | 0.1074 | No |
| 41 | Btg1 | 5059 | 0.184 | 0.1111 | No |
| 42 | Jun | 5099 | 0.182 | 0.1133 | No |
| 43 | Ampd3 | 5110 | 0.182 | 0.1170 | No |
| 44 | Fosl2 | 5240 | 0.175 | 0.1146 | No |
| 45 | Chst3 | 5313 | 0.173 | 0.1149 | No |
| 46 | Rora | 5386 | 0.169 | 0.1152 | No |
| 47 | Col5a1 | 5405 | 0.168 | 0.1182 | No |
| 48 | Tpi1 | 5439 | 0.166 | 0.1204 | No |
| 49 | Ddit4 | 5473 | 0.164 | 0.1225 | No |
| 50 | Akap12 | 5518 | 0.162 | 0.1240 | No |
| 51 | Cdkn1a | 5553 | 0.161 | 0.1260 | No |
| 52 | Pfkfb3 | 5694 | 0.154 | 0.1225 | No |
| 53 | Vhl | 5719 | 0.153 | 0.1248 | No |
| 54 | Klhl24 | 5870 | 0.146 | 0.1207 | No |
| 55 | Zfp292 | 5948 | 0.143 | 0.1201 | No |
| 56 | B4galnt2 | 6051 | 0.138 | 0.1181 | No |
| 57 | Zfp36 | 6054 | 0.138 | 0.1212 | No |
| 58 | Map3k1 | 6979 | 0.101 | 0.0770 | No |
| 59 | Klf7 | 7001 | 0.100 | 0.0783 | No |
| 60 | Car12 | 7139 | 0.094 | 0.0736 | No |
| 61 | Isg20 | 7443 | 0.081 | 0.0602 | No |
| 62 | Mt1 | 7574 | 0.075 | 0.0554 | No |
| 63 | Vegfa | 7594 | 0.074 | 0.0561 | No |
| 64 | Gpc3 | 7679 | 0.070 | 0.0535 | No |
| 65 | Gaa | 7945 | 0.059 | 0.0416 | No |
| 66 | Pgk1 | 7995 | 0.058 | 0.0404 | No |
| 67 | Ldhc | 8271 | 0.045 | 0.0276 | No |
| 68 | Xpnpep1 | 8363 | 0.041 | 0.0240 | No |
| 69 | Sdc3 | 8469 | 0.037 | 0.0196 | No |
| 70 | Atf3 | 8529 | 0.035 | 0.0174 | No |
| 71 | Nfil3 | 8897 | 0.017 | -0.0007 | No |
| 72 | Mxi1 | 8908 | 0.017 | -0.0008 | No |
| 73 | Pgam2 | 8953 | 0.015 | -0.0027 | No |
| 74 | Gpc4 | 9024 | 0.012 | -0.0059 | No |
| 75 | Ldha | 9285 | 0.000 | -0.0190 | No |
| 76 | Ak4 | 9393 | -0.003 | -0.0243 | No |
| 77 | Pfkl | 9597 | -0.012 | -0.0343 | No |
| 78 | Scarb1 | 9656 | -0.015 | -0.0368 | No |
| 79 | Ccng2 | 9664 | -0.015 | -0.0369 | No |
| 80 | Tes | 9714 | -0.018 | -0.0389 | No |
| 81 | Gbe1 | 9826 | -0.022 | -0.0440 | No |
| 82 | Pdk3 | 9832 | -0.022 | -0.0437 | No |
| 83 | Slc2a5 | 9907 | -0.024 | -0.0469 | No |
| 84 | Plaur | 9953 | -0.026 | -0.0486 | No |
| 85 | Wsb1 | 10080 | -0.032 | -0.0542 | No |
| 86 | Kdm3a | 10096 | -0.033 | -0.0542 | No |
| 87 | Kif5a | 10248 | -0.040 | -0.0609 | No |
| 88 | Ankzf1 | 10450 | -0.048 | -0.0699 | No |
| 89 | Ndst2 | 10957 | -0.069 | -0.0938 | No |
| 90 | Pdk1 | 10981 | -0.070 | -0.0933 | No |
| 91 | Slc37a4 | 11086 | -0.072 | -0.0969 | No |
| 92 | Csrp2 | 11180 | -0.076 | -0.0998 | No |
| 93 | Nedd4l | 11203 | -0.077 | -0.0991 | No |
| 94 | Pygm | 11244 | -0.079 | -0.0993 | No |
| 95 | Stc2 | 11462 | -0.088 | -0.1082 | No |
| 96 | Hk1 | 11764 | -0.101 | -0.1210 | No |
| 97 | Mif | 11887 | -0.107 | -0.1247 | No |
| 98 | Ets1 | 11967 | -0.110 | -0.1261 | No |
| 99 | Tpst2 | 11971 | -0.111 | -0.1237 | No |
| 100 | Fam162a | 12089 | -0.116 | -0.1269 | No |
| 101 | Hs3st1 | 12375 | -0.127 | -0.1383 | No |
| 102 | Plac8 | 12441 | -0.131 | -0.1386 | No |
| 103 | Tgfbi | 12481 | -0.133 | -0.1375 | No |
| 104 | Hk2 | 12519 | -0.136 | -0.1362 | No |
| 105 | Pgm1 | 12638 | -0.141 | -0.1389 | No |
| 106 | Pklr | 12782 | -0.147 | -0.1427 | No |
| 107 | Dtna | 12787 | -0.148 | -0.1395 | No |
| 108 | Dcn | 12946 | -0.155 | -0.1439 | No |
| 109 | Rbpj | 13010 | -0.157 | -0.1434 | No |
| 110 | Hexa | 13279 | -0.168 | -0.1530 | No |
| 111 | Slc2a1 | 13459 | -0.178 | -0.1579 | No |
| 112 | Ndst1 | 13467 | -0.178 | -0.1542 | No |
| 113 | Galk1 | 13784 | -0.192 | -0.1656 | No |
| 114 | Eno3 | 13963 | -0.200 | -0.1700 | No |
| 115 | Slc25a1 | 14025 | -0.203 | -0.1684 | No |
| 116 | Pam | 14274 | -0.217 | -0.1759 | No |
| 117 | Cavin1 | 14435 | -0.225 | -0.1787 | No |
| 118 | Aldoc | 14502 | -0.228 | -0.1768 | No |
| 119 | Gys1 | 14552 | -0.230 | -0.1739 | No |
| 120 | Ppp1r15a | 14617 | -0.233 | -0.1718 | No |
| 121 | Lxn | 14766 | -0.242 | -0.1736 | No |
| 122 | Gcnt2 | 14804 | -0.244 | -0.1699 | No |
| 123 | Casp6 | 14824 | -0.245 | -0.1652 | No |
| 124 | Bnip3l | 14829 | -0.245 | -0.1597 | No |
| 125 | Cp | 14848 | -0.246 | -0.1550 | No |
| 126 | Ids | 15053 | -0.254 | -0.1594 | No |
| 127 | Eno1b | 15521 | -0.279 | -0.1764 | No |
| 128 | Vldlr | 15624 | -0.285 | -0.1750 | No |
| 129 | Tnfaip3 | 15746 | -0.291 | -0.1744 | No |
| 130 | Adora2b | 15748 | -0.291 | -0.1677 | No |
| 131 | Ext1 | 15798 | -0.294 | -0.1634 | No |
| 132 | B3galt6 | 15826 | -0.296 | -0.1579 | No |
| 133 | Cav1 | 15866 | -0.298 | -0.1530 | No |
| 134 | Gpc1 | 16152 | -0.314 | -0.1601 | No |
| 135 | Glrx | 16429 | -0.329 | -0.1664 | No |
| 136 | Grhpr | 16702 | -0.345 | -0.1721 | No |
| 137 | Tgm2 | 16879 | -0.357 | -0.1727 | No |
| 138 | Siah2 | 17187 | -0.377 | -0.1795 | No |
| 139 | Cxcr4 | 17610 | -0.407 | -0.1914 | Yes |
| 140 | Kdelr3 | 17735 | -0.419 | -0.1879 | Yes |
| 141 | Eno2 | 17864 | -0.431 | -0.1844 | Yes |
| 142 | Tmem45a | 17902 | -0.434 | -0.1763 | Yes |
| 143 | Errfi1 | 17961 | -0.440 | -0.1690 | Yes |
| 144 | Prkca | 18274 | -0.469 | -0.1739 | Yes |
| 145 | F3 | 18696 | -0.516 | -0.1832 | Yes |
| 146 | Atp7a | 18782 | -0.529 | -0.1753 | Yes |
| 147 | Sult2b1 | 18838 | -0.538 | -0.1657 | Yes |
| 148 | Ddit3 | 18849 | -0.539 | -0.1537 | Yes |
| 149 | Nagk | 18853 | -0.539 | -0.1414 | Yes |
| 150 | Cited2 | 18873 | -0.541 | -0.1299 | Yes |
| 151 | Fbp1 | 18876 | -0.541 | -0.1175 | Yes |
| 152 | Ppfia4 | 19041 | -0.567 | -0.1127 | Yes |
| 153 | Gapdhs | 19185 | -0.593 | -0.1062 | Yes |
| 154 | Bcl2 | 19197 | -0.596 | -0.0930 | Yes |
| 155 | Slc2a3 | 19280 | -0.613 | -0.0830 | Yes |
| 156 | Selenbp1 | 19330 | -0.627 | -0.0710 | Yes |
| 157 | Pnrc1 | 19401 | -0.644 | -0.0596 | Yes |
| 158 | Sdc2 | 19503 | -0.670 | -0.0493 | Yes |
| 159 | Angptl4 | 19515 | -0.673 | -0.0343 | Yes |
| 160 | Ier3 | 19736 | -0.767 | -0.0277 | Yes |
| 161 | Prdx5 | 19749 | -0.777 | -0.0103 | Yes |
| 162 | Ilvbl | 19978 | -1.050 | 0.0024 | Yes |