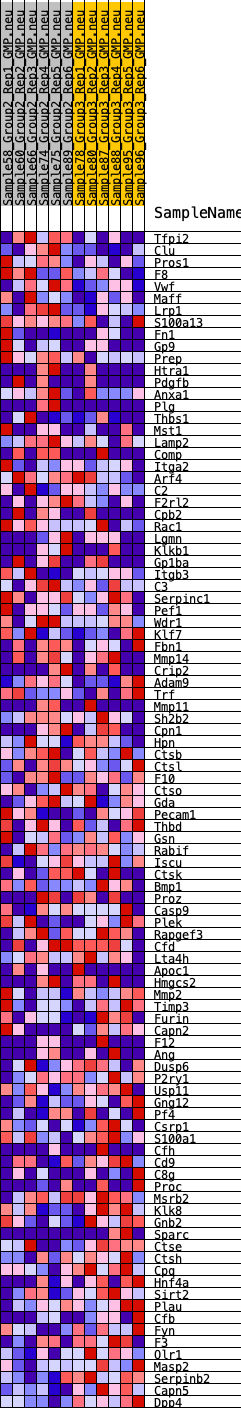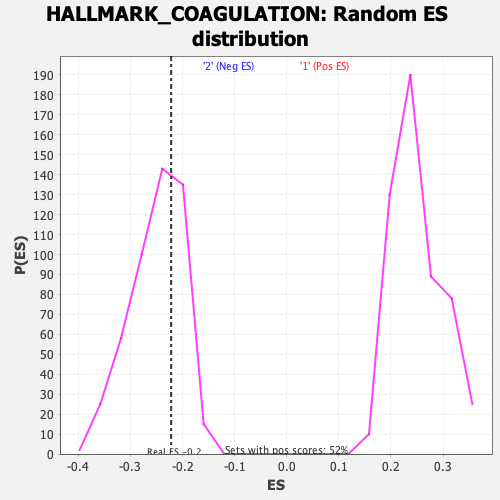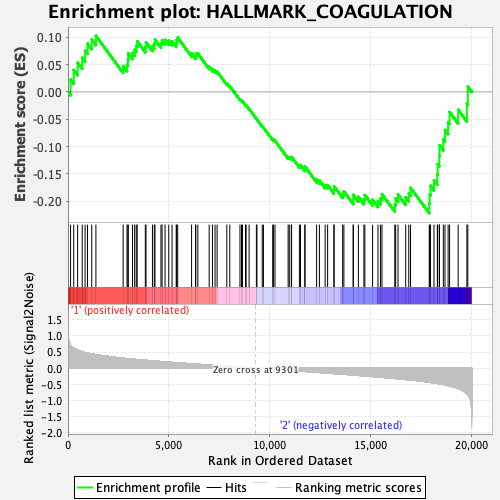
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | GMP.GMP.neu_Pheno.cls #Group2_versus_Group3.GMP.neu_Pheno.cls #Group2_versus_Group3_repos |
| Phenotype | GMP.neu_Pheno.cls#Group2_versus_Group3_repos |
| Upregulated in class | 2 |
| GeneSet | HALLMARK_COAGULATION |
| Enrichment Score (ES) | -0.22179852 |
| Normalized Enrichment Score (NES) | -0.8826261 |
| Nominal p-value | 0.667364 |
| FDR q-value | 0.7842102 |
| FWER p-Value | 0.997 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Tfpi2 | 127 | 0.715 | 0.0226 | No |
| 2 | Clu | 284 | 0.628 | 0.0403 | No |
| 3 | Pros1 | 478 | 0.566 | 0.0536 | No |
| 4 | F8 | 705 | 0.510 | 0.0629 | No |
| 5 | Vwf | 848 | 0.487 | 0.0755 | No |
| 6 | Maff | 968 | 0.469 | 0.0886 | No |
| 7 | Lrp1 | 1178 | 0.443 | 0.0961 | No |
| 8 | S100a13 | 1384 | 0.417 | 0.1027 | No |
| 9 | Fn1 | 2735 | 0.309 | 0.0475 | No |
| 10 | Gp9 | 2927 | 0.296 | 0.0499 | No |
| 11 | Prep | 2974 | 0.294 | 0.0595 | No |
| 12 | Htra1 | 2987 | 0.293 | 0.0708 | No |
| 13 | Pdgfb | 3201 | 0.279 | 0.0715 | No |
| 14 | Anxa1 | 3306 | 0.274 | 0.0773 | No |
| 15 | Plg | 3384 | 0.270 | 0.0844 | No |
| 16 | Thbs1 | 3431 | 0.268 | 0.0930 | No |
| 17 | Mst1 | 3834 | 0.245 | 0.0827 | No |
| 18 | Lamp2 | 3869 | 0.244 | 0.0909 | No |
| 19 | Comp | 4188 | 0.227 | 0.0842 | No |
| 20 | Itga2 | 4279 | 0.222 | 0.0887 | No |
| 21 | Arf4 | 4313 | 0.219 | 0.0959 | No |
| 22 | C2 | 4611 | 0.206 | 0.0893 | No |
| 23 | F2rl2 | 4665 | 0.201 | 0.0949 | No |
| 24 | Cpb2 | 4813 | 0.193 | 0.0953 | No |
| 25 | Rac1 | 4996 | 0.187 | 0.0938 | No |
| 26 | Lgmn | 5160 | 0.179 | 0.0929 | No |
| 27 | Klkb1 | 5366 | 0.170 | 0.0895 | No |
| 28 | Gp1ba | 5387 | 0.169 | 0.0953 | No |
| 29 | Itgb3 | 5440 | 0.166 | 0.0995 | No |
| 30 | C3 | 6129 | 0.135 | 0.0704 | No |
| 31 | Serpinc1 | 6329 | 0.126 | 0.0656 | No |
| 32 | Pef1 | 6332 | 0.126 | 0.0706 | No |
| 33 | Wdr1 | 6438 | 0.122 | 0.0703 | No |
| 34 | Klf7 | 7001 | 0.100 | 0.0461 | No |
| 35 | Fbn1 | 7165 | 0.092 | 0.0417 | No |
| 36 | Mmp14 | 7292 | 0.089 | 0.0390 | No |
| 37 | Crip2 | 7395 | 0.084 | 0.0373 | No |
| 38 | Adam9 | 7875 | 0.062 | 0.0158 | No |
| 39 | Trf | 8024 | 0.057 | 0.0106 | No |
| 40 | Mmp11 | 8528 | 0.035 | -0.0132 | No |
| 41 | Sh2b2 | 8606 | 0.031 | -0.0158 | No |
| 42 | Cpn1 | 8656 | 0.028 | -0.0171 | No |
| 43 | Hpn | 8802 | 0.021 | -0.0235 | No |
| 44 | Ctsb | 8834 | 0.020 | -0.0243 | No |
| 45 | Ctsl | 8981 | 0.013 | -0.0311 | No |
| 46 | F10 | 9342 | -0.001 | -0.0491 | No |
| 47 | Ctso | 9369 | -0.002 | -0.0503 | No |
| 48 | Gda | 9632 | -0.014 | -0.0629 | No |
| 49 | Pecam1 | 9693 | -0.016 | -0.0653 | No |
| 50 | Thbd | 10153 | -0.035 | -0.0869 | No |
| 51 | Gsn | 10186 | -0.037 | -0.0870 | No |
| 52 | Rabif | 10265 | -0.040 | -0.0892 | No |
| 53 | Iscu | 10917 | -0.067 | -0.1192 | No |
| 54 | Ctsk | 10974 | -0.069 | -0.1192 | No |
| 55 | Bmp1 | 11074 | -0.072 | -0.1212 | No |
| 56 | Proz | 11088 | -0.072 | -0.1189 | No |
| 57 | Casp9 | 11475 | -0.089 | -0.1347 | No |
| 58 | Plek | 11530 | -0.091 | -0.1337 | No |
| 59 | Rapgef3 | 11749 | -0.100 | -0.1406 | No |
| 60 | Cfd | 11750 | -0.100 | -0.1365 | No |
| 61 | Lta4h | 12330 | -0.125 | -0.1605 | No |
| 62 | Apoc1 | 12468 | -0.132 | -0.1620 | No |
| 63 | Hmgcs2 | 12752 | -0.146 | -0.1703 | No |
| 64 | Mmp2 | 12875 | -0.151 | -0.1703 | No |
| 65 | Timp3 | 13187 | -0.165 | -0.1792 | No |
| 66 | Furin | 13195 | -0.165 | -0.1729 | No |
| 67 | Capn2 | 13620 | -0.185 | -0.1866 | No |
| 68 | F12 | 13682 | -0.188 | -0.1821 | No |
| 69 | Ang | 14144 | -0.209 | -0.1967 | No |
| 70 | Dusp6 | 14147 | -0.209 | -0.1883 | No |
| 71 | P2ry1 | 14400 | -0.224 | -0.1919 | No |
| 72 | Usp11 | 14675 | -0.236 | -0.1960 | No |
| 73 | Gng12 | 14722 | -0.239 | -0.1886 | No |
| 74 | Pf4 | 15106 | -0.258 | -0.1974 | No |
| 75 | Csrp1 | 15372 | -0.271 | -0.1997 | No |
| 76 | S100a1 | 15496 | -0.277 | -0.1947 | No |
| 77 | Cfh | 15571 | -0.282 | -0.1870 | No |
| 78 | Cd9 | 16206 | -0.318 | -0.2059 | Yes |
| 79 | C8g | 16250 | -0.320 | -0.1950 | Yes |
| 80 | Proc | 16367 | -0.326 | -0.1876 | Yes |
| 81 | Msrb2 | 16748 | -0.347 | -0.1926 | Yes |
| 82 | Klk8 | 16900 | -0.357 | -0.1857 | Yes |
| 83 | Gnb2 | 16987 | -0.363 | -0.1753 | Yes |
| 84 | Sparc | 17915 | -0.435 | -0.2041 | Yes |
| 85 | Ctse | 17935 | -0.437 | -0.1874 | Yes |
| 86 | Ctsh | 17979 | -0.441 | -0.1716 | Yes |
| 87 | Cpq | 18150 | -0.456 | -0.1616 | Yes |
| 88 | Hnf4a | 18316 | -0.473 | -0.1507 | Yes |
| 89 | Sirt2 | 18332 | -0.475 | -0.1322 | Yes |
| 90 | Plau | 18414 | -0.484 | -0.1166 | Yes |
| 91 | Cfb | 18424 | -0.485 | -0.0974 | Yes |
| 92 | Fyn | 18614 | -0.506 | -0.0864 | Yes |
| 93 | F3 | 18696 | -0.516 | -0.0695 | Yes |
| 94 | Olr1 | 18851 | -0.539 | -0.0554 | Yes |
| 95 | Masp2 | 18924 | -0.549 | -0.0367 | Yes |
| 96 | Serpinb2 | 19349 | -0.631 | -0.0324 | Yes |
| 97 | Capn5 | 19782 | -0.803 | -0.0215 | Yes |
| 98 | Dpp4 | 19828 | -0.830 | 0.0099 | Yes |