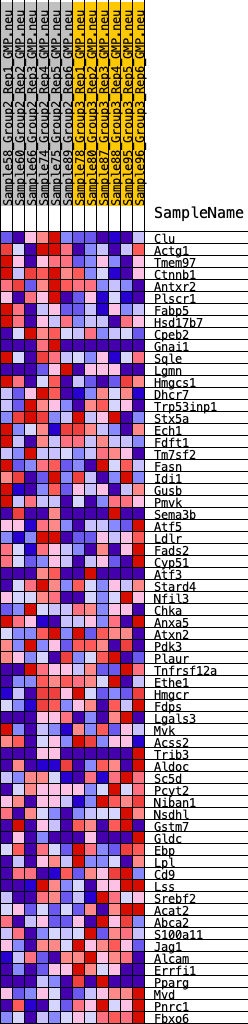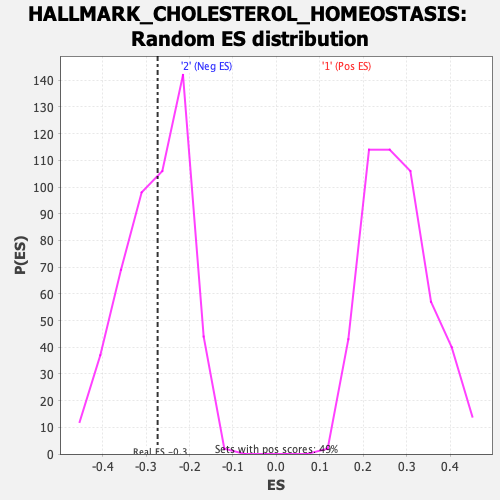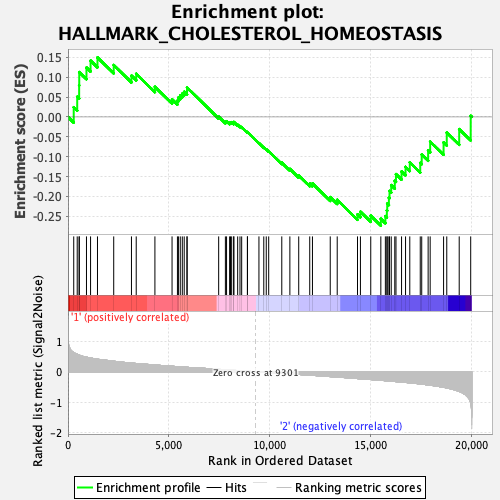
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | GMP.GMP.neu_Pheno.cls #Group2_versus_Group3.GMP.neu_Pheno.cls #Group2_versus_Group3_repos |
| Phenotype | GMP.neu_Pheno.cls#Group2_versus_Group3_repos |
| Upregulated in class | 2 |
| GeneSet | HALLMARK_CHOLESTEROL_HOMEOSTASIS |
| Enrichment Score (ES) | -0.2734806 |
| Normalized Enrichment Score (NES) | -0.98592806 |
| Nominal p-value | 0.4627451 |
| FDR q-value | 0.80292505 |
| FWER p-Value | 0.989 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Clu | 284 | 0.628 | 0.0244 | No |
| 2 | Actg1 | 457 | 0.570 | 0.0508 | No |
| 3 | Tmem97 | 553 | 0.545 | 0.0796 | No |
| 4 | Ctnnb1 | 558 | 0.544 | 0.1128 | No |
| 5 | Antxr2 | 916 | 0.477 | 0.1243 | No |
| 6 | Plscr1 | 1119 | 0.448 | 0.1417 | No |
| 7 | Fabp5 | 1462 | 0.410 | 0.1498 | No |
| 8 | Hsd17b7 | 2267 | 0.343 | 0.1307 | No |
| 9 | Cpeb2 | 3147 | 0.282 | 0.1039 | No |
| 10 | Gnai1 | 3383 | 0.270 | 0.1087 | No |
| 11 | Sqle | 4310 | 0.220 | 0.0759 | No |
| 12 | Lgmn | 5160 | 0.179 | 0.0444 | No |
| 13 | Hmgcs1 | 5427 | 0.167 | 0.0413 | No |
| 14 | Dhcr7 | 5476 | 0.164 | 0.0490 | No |
| 15 | Trp53inp1 | 5567 | 0.160 | 0.0543 | No |
| 16 | Stx5a | 5668 | 0.156 | 0.0589 | No |
| 17 | Ech1 | 5763 | 0.151 | 0.0634 | No |
| 18 | Fdft1 | 5902 | 0.145 | 0.0654 | No |
| 19 | Tm7sf2 | 5905 | 0.145 | 0.0742 | No |
| 20 | Fasn | 7472 | 0.080 | 0.0007 | No |
| 21 | Idi1 | 7805 | 0.066 | -0.0119 | No |
| 22 | Gusb | 7863 | 0.063 | -0.0109 | No |
| 23 | Pmvk | 8015 | 0.057 | -0.0150 | No |
| 24 | Sema3b | 8059 | 0.055 | -0.0137 | No |
| 25 | Atf5 | 8116 | 0.053 | -0.0133 | No |
| 26 | Ldlr | 8219 | 0.048 | -0.0155 | No |
| 27 | Fads2 | 8223 | 0.047 | -0.0127 | No |
| 28 | Cyp51 | 8414 | 0.039 | -0.0198 | No |
| 29 | Atf3 | 8529 | 0.035 | -0.0234 | No |
| 30 | Stard4 | 8613 | 0.030 | -0.0257 | No |
| 31 | Nfil3 | 8897 | 0.017 | -0.0388 | No |
| 32 | Chka | 8900 | 0.017 | -0.0379 | No |
| 33 | Anxa5 | 9469 | -0.006 | -0.0659 | No |
| 34 | Atxn2 | 9709 | -0.017 | -0.0768 | No |
| 35 | Pdk3 | 9832 | -0.022 | -0.0816 | No |
| 36 | Plaur | 9953 | -0.026 | -0.0860 | No |
| 37 | Tnfrsf12a | 10602 | -0.054 | -0.1151 | No |
| 38 | Ethe1 | 11003 | -0.071 | -0.1308 | No |
| 39 | Hmgcr | 11442 | -0.087 | -0.1474 | No |
| 40 | Fdps | 11988 | -0.112 | -0.1679 | No |
| 41 | Lgals3 | 12123 | -0.118 | -0.1673 | No |
| 42 | Mvk | 13005 | -0.157 | -0.2018 | No |
| 43 | Acss2 | 13351 | -0.172 | -0.2085 | No |
| 44 | Trib3 | 14357 | -0.221 | -0.2453 | No |
| 45 | Aldoc | 14502 | -0.228 | -0.2384 | No |
| 46 | Sc5d | 15014 | -0.253 | -0.2485 | No |
| 47 | Pcyt2 | 15514 | -0.278 | -0.2564 | Yes |
| 48 | Niban1 | 15739 | -0.291 | -0.2497 | Yes |
| 49 | Nsdhl | 15813 | -0.295 | -0.2352 | Yes |
| 50 | Gstm7 | 15832 | -0.296 | -0.2179 | Yes |
| 51 | Gldc | 15914 | -0.301 | -0.2035 | Yes |
| 52 | Ebp | 15945 | -0.303 | -0.1863 | Yes |
| 53 | Lpl | 16030 | -0.308 | -0.1716 | Yes |
| 54 | Cd9 | 16206 | -0.318 | -0.1608 | Yes |
| 55 | Lss | 16265 | -0.321 | -0.1440 | Yes |
| 56 | Srebf2 | 16542 | -0.335 | -0.1372 | Yes |
| 57 | Acat2 | 16740 | -0.347 | -0.1258 | Yes |
| 58 | Abca2 | 16950 | -0.360 | -0.1141 | Yes |
| 59 | S100a11 | 17469 | -0.397 | -0.1156 | Yes |
| 60 | Jag1 | 17542 | -0.402 | -0.0945 | Yes |
| 61 | Alcam | 17860 | -0.430 | -0.0839 | Yes |
| 62 | Errfi1 | 17961 | -0.440 | -0.0619 | Yes |
| 63 | Pparg | 18626 | -0.507 | -0.0640 | Yes |
| 64 | Mvd | 18784 | -0.529 | -0.0394 | Yes |
| 65 | Pnrc1 | 19401 | -0.644 | -0.0307 | Yes |
| 66 | Fbxo6 | 19971 | -1.008 | 0.0028 | Yes |