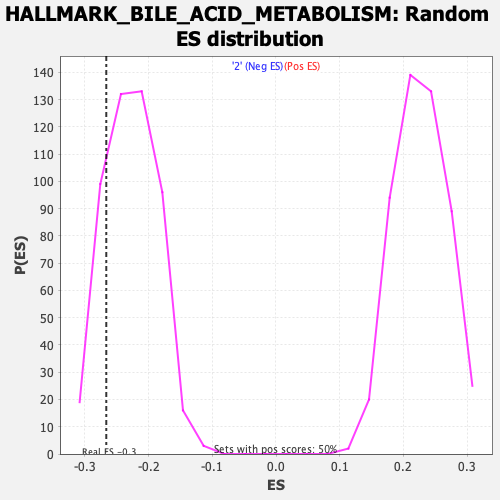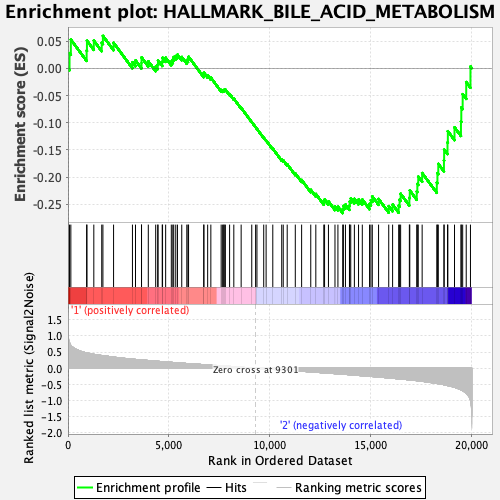
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | GMP.GMP.neu_Pheno.cls #Group2_versus_Group3.GMP.neu_Pheno.cls #Group2_versus_Group3_repos |
| Phenotype | GMP.neu_Pheno.cls#Group2_versus_Group3_repos |
| Upregulated in class | 2 |
| GeneSet | HALLMARK_BILE_ACID_METABOLISM |
| Enrichment Score (ES) | -0.2664803 |
| Normalized Enrichment Score (NES) | -1.175924 |
| Nominal p-value | 0.16064256 |
| FDR q-value | 1.0 |
| FWER p-Value | 0.915 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Abca1 | 76 | 0.792 | 0.0279 | No |
| 2 | Gclm | 141 | 0.704 | 0.0528 | No |
| 3 | Bcar3 | 924 | 0.476 | 0.0326 | No |
| 4 | Pfkm | 940 | 0.474 | 0.0508 | No |
| 5 | Cyp46a1 | 1280 | 0.430 | 0.0510 | No |
| 6 | Abcg4 | 1673 | 0.393 | 0.0471 | No |
| 7 | Hao1 | 1731 | 0.387 | 0.0597 | No |
| 8 | Scp2 | 2260 | 0.344 | 0.0470 | No |
| 9 | Lck | 3196 | 0.280 | 0.0112 | No |
| 10 | Abca5 | 3347 | 0.272 | 0.0146 | No |
| 11 | Fads1 | 3645 | 0.256 | 0.0099 | No |
| 12 | Aldh9a1 | 3647 | 0.256 | 0.0201 | No |
| 13 | Nr1h4 | 3982 | 0.239 | 0.0129 | No |
| 14 | Abca3 | 4349 | 0.218 | 0.0032 | No |
| 15 | Dio2 | 4450 | 0.213 | 0.0067 | No |
| 16 | Idh1 | 4465 | 0.212 | 0.0145 | No |
| 17 | Acsl5 | 4673 | 0.201 | 0.0122 | No |
| 18 | Gnmt | 4689 | 0.200 | 0.0195 | No |
| 19 | Retsat | 4843 | 0.192 | 0.0195 | No |
| 20 | Aqp9 | 5123 | 0.181 | 0.0127 | No |
| 21 | Efhc1 | 5198 | 0.177 | 0.0161 | No |
| 22 | Pex6 | 5244 | 0.175 | 0.0208 | No |
| 23 | Slc23a2 | 5344 | 0.171 | 0.0227 | No |
| 24 | Hsd17b4 | 5428 | 0.167 | 0.0252 | No |
| 25 | Slc35b2 | 5641 | 0.157 | 0.0209 | No |
| 26 | Aldh8a1 | 5892 | 0.145 | 0.0142 | No |
| 27 | Sod1 | 5950 | 0.143 | 0.0170 | No |
| 28 | Paox | 5979 | 0.141 | 0.0212 | No |
| 29 | Slc22a18 | 6732 | 0.111 | -0.0121 | No |
| 30 | Optn | 6740 | 0.110 | -0.0080 | No |
| 31 | Ephx2 | 6926 | 0.104 | -0.0131 | No |
| 32 | Fdxr | 7084 | 0.097 | -0.0171 | No |
| 33 | Lonp2 | 7596 | 0.074 | -0.0398 | No |
| 34 | Dhcr24 | 7669 | 0.071 | -0.0406 | No |
| 35 | Crot | 7720 | 0.069 | -0.0404 | No |
| 36 | Gnpat | 7751 | 0.068 | -0.0391 | No |
| 37 | Idi1 | 7805 | 0.066 | -0.0392 | No |
| 38 | Abcd3 | 8014 | 0.057 | -0.0473 | No |
| 39 | Fads2 | 8223 | 0.047 | -0.0559 | No |
| 40 | Abcd1 | 8587 | 0.032 | -0.0728 | No |
| 41 | Abca8b | 9113 | 0.007 | -0.0989 | No |
| 42 | Gc | 9304 | 0.000 | -0.1084 | No |
| 43 | Npc1 | 9370 | -0.002 | -0.1116 | No |
| 44 | Soat2 | 9706 | -0.017 | -0.1277 | No |
| 45 | Gstk1 | 9825 | -0.022 | -0.1327 | No |
| 46 | Pex16 | 10156 | -0.035 | -0.1479 | No |
| 47 | Slc27a5 | 10599 | -0.054 | -0.1679 | No |
| 48 | Pecr | 10674 | -0.056 | -0.1693 | No |
| 49 | Pxmp2 | 10871 | -0.065 | -0.1766 | No |
| 50 | Abcd2 | 11272 | -0.080 | -0.1934 | No |
| 51 | Acsl1 | 11586 | -0.093 | -0.2054 | No |
| 52 | Pex11g | 12039 | -0.114 | -0.2235 | No |
| 53 | Atxn1 | 12290 | -0.123 | -0.2312 | No |
| 54 | Cyp7b1 | 12686 | -0.143 | -0.2452 | No |
| 55 | Pnpla8 | 12724 | -0.144 | -0.2413 | No |
| 56 | Pex13 | 12916 | -0.154 | -0.2448 | No |
| 57 | Pex19 | 13240 | -0.167 | -0.2543 | No |
| 58 | Cyp27a1 | 13386 | -0.174 | -0.2546 | No |
| 59 | Cat | 13624 | -0.185 | -0.2591 | Yes |
| 60 | Amacr | 13657 | -0.187 | -0.2532 | Yes |
| 61 | Isoc1 | 13759 | -0.192 | -0.2506 | Yes |
| 62 | Aldh1a1 | 13958 | -0.200 | -0.2525 | Yes |
| 63 | Slc29a1 | 13971 | -0.200 | -0.2451 | Yes |
| 64 | Nedd4 | 14022 | -0.203 | -0.2395 | Yes |
| 65 | Pex26 | 14202 | -0.213 | -0.2400 | Yes |
| 66 | Pex7 | 14409 | -0.224 | -0.2414 | Yes |
| 67 | Rxra | 14595 | -0.231 | -0.2414 | Yes |
| 68 | Abca6 | 14955 | -0.250 | -0.2494 | Yes |
| 69 | Akr1d1 | 15022 | -0.253 | -0.2425 | Yes |
| 70 | Pex11a | 15091 | -0.257 | -0.2357 | Yes |
| 71 | Nr3c2 | 15402 | -0.272 | -0.2404 | Yes |
| 72 | Slc27a2 | 15906 | -0.301 | -0.2536 | Yes |
| 73 | Abca4 | 16098 | -0.313 | -0.2506 | Yes |
| 74 | Hacl1 | 16399 | -0.328 | -0.2526 | Yes |
| 75 | Idh2 | 16444 | -0.330 | -0.2416 | Yes |
| 76 | Hsd17b11 | 16496 | -0.332 | -0.2308 | Yes |
| 77 | Hsd3b7 | 16929 | -0.360 | -0.2381 | Yes |
| 78 | Abca2 | 16950 | -0.360 | -0.2247 | Yes |
| 79 | Ar | 17295 | -0.384 | -0.2266 | Yes |
| 80 | Rbp1 | 17325 | -0.386 | -0.2126 | Yes |
| 81 | Cyp39a1 | 17370 | -0.390 | -0.1992 | Yes |
| 82 | Phyh | 17563 | -0.404 | -0.1927 | Yes |
| 83 | Pex1 | 18290 | -0.470 | -0.2103 | Yes |
| 84 | Bmp6 | 18321 | -0.474 | -0.1928 | Yes |
| 85 | Slc23a1 | 18364 | -0.478 | -0.1758 | Yes |
| 86 | Nr0b2 | 18644 | -0.509 | -0.1694 | Yes |
| 87 | Pex12 | 18654 | -0.510 | -0.1495 | Yes |
| 88 | Tfcp2l1 | 18821 | -0.535 | -0.1364 | Yes |
| 89 | Sult2b1 | 18838 | -0.538 | -0.1157 | Yes |
| 90 | Klf1 | 19168 | -0.591 | -0.1086 | Yes |
| 91 | Nudt12 | 19488 | -0.666 | -0.0979 | Yes |
| 92 | Lipe | 19505 | -0.670 | -0.0719 | Yes |
| 93 | Mlycd | 19573 | -0.692 | -0.0476 | Yes |
| 94 | Prdx5 | 19749 | -0.777 | -0.0253 | Yes |
| 95 | Abca9 | 19959 | -0.978 | 0.0034 | Yes |