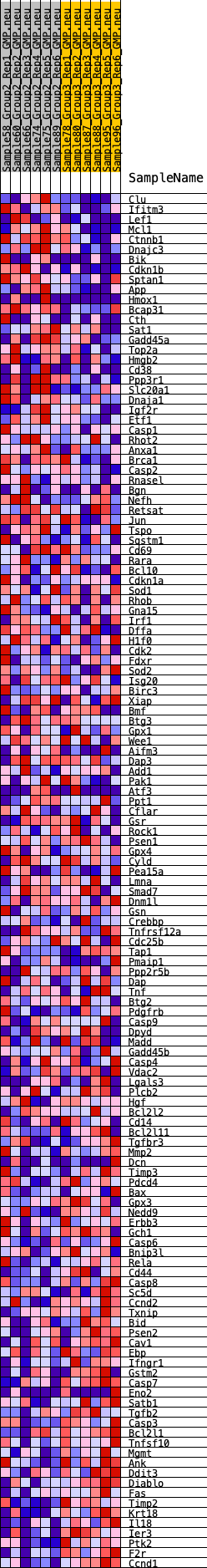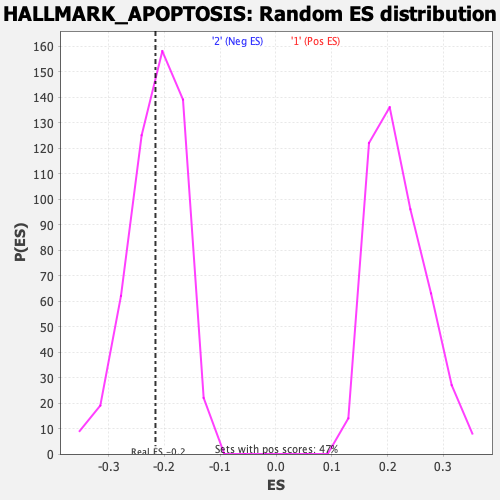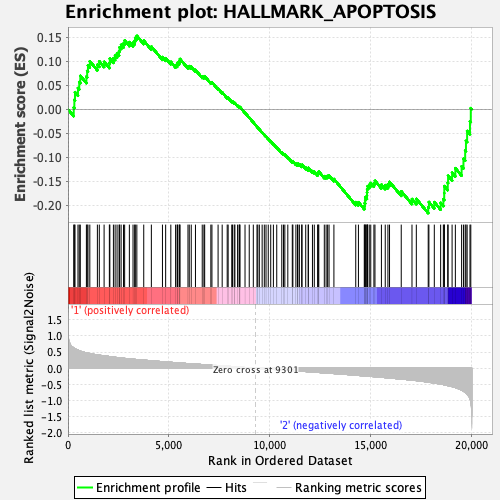
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | GMP.GMP.neu_Pheno.cls #Group2_versus_Group3.GMP.neu_Pheno.cls #Group2_versus_Group3_repos |
| Phenotype | GMP.neu_Pheno.cls#Group2_versus_Group3_repos |
| Upregulated in class | 2 |
| GeneSet | HALLMARK_APOPTOSIS |
| Enrichment Score (ES) | -0.21614204 |
| Normalized Enrichment Score (NES) | -1.0036745 |
| Nominal p-value | 0.4681648 |
| FDR q-value | 0.87112474 |
| FWER p-Value | 0.986 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Clu | 284 | 0.628 | 0.0038 | No |
| 2 | Ifitm3 | 315 | 0.612 | 0.0199 | No |
| 3 | Lef1 | 347 | 0.601 | 0.0357 | No |
| 4 | Mcl1 | 495 | 0.562 | 0.0445 | No |
| 5 | Ctnnb1 | 558 | 0.544 | 0.0570 | No |
| 6 | Dnajc3 | 610 | 0.530 | 0.0697 | No |
| 7 | Bik | 912 | 0.477 | 0.0683 | No |
| 8 | Cdkn1b | 951 | 0.472 | 0.0800 | No |
| 9 | Sptan1 | 990 | 0.466 | 0.0915 | No |
| 10 | App | 1082 | 0.454 | 0.1000 | No |
| 11 | Hmox1 | 1457 | 0.411 | 0.0930 | No |
| 12 | Bcap31 | 1552 | 0.402 | 0.0998 | No |
| 13 | Cth | 1790 | 0.382 | 0.0989 | No |
| 14 | Sat1 | 2051 | 0.361 | 0.0962 | No |
| 15 | Gadd45a | 2075 | 0.360 | 0.1054 | No |
| 16 | Top2a | 2252 | 0.345 | 0.1065 | No |
| 17 | Hmgb2 | 2336 | 0.339 | 0.1121 | No |
| 18 | Cd38 | 2433 | 0.330 | 0.1168 | No |
| 19 | Ppp3r1 | 2532 | 0.322 | 0.1211 | No |
| 20 | Slc20a1 | 2558 | 0.321 | 0.1291 | No |
| 21 | Dnaja1 | 2634 | 0.315 | 0.1344 | No |
| 22 | Igf2r | 2753 | 0.308 | 0.1373 | No |
| 23 | Etf1 | 2809 | 0.304 | 0.1433 | No |
| 24 | Casp1 | 3043 | 0.290 | 0.1399 | No |
| 25 | Rhot2 | 3225 | 0.278 | 0.1388 | No |
| 26 | Anxa1 | 3306 | 0.274 | 0.1427 | No |
| 27 | Brca1 | 3338 | 0.272 | 0.1490 | No |
| 28 | Casp2 | 3414 | 0.268 | 0.1529 | No |
| 29 | Rnasel | 3756 | 0.249 | 0.1430 | No |
| 30 | Bgn | 4134 | 0.230 | 0.1306 | No |
| 31 | Nefh | 4684 | 0.201 | 0.1088 | No |
| 32 | Retsat | 4843 | 0.192 | 0.1064 | No |
| 33 | Jun | 5099 | 0.182 | 0.0988 | No |
| 34 | Tspo | 5336 | 0.172 | 0.0919 | No |
| 35 | Sqstm1 | 5411 | 0.168 | 0.0930 | No |
| 36 | Cd69 | 5432 | 0.167 | 0.0968 | No |
| 37 | Rara | 5509 | 0.162 | 0.0976 | No |
| 38 | Bcl10 | 5538 | 0.162 | 0.1009 | No |
| 39 | Cdkn1a | 5553 | 0.161 | 0.1048 | No |
| 40 | Sod1 | 5950 | 0.143 | 0.0890 | No |
| 41 | Rhob | 6013 | 0.140 | 0.0899 | No |
| 42 | Gna15 | 6112 | 0.135 | 0.0889 | No |
| 43 | Irf1 | 6321 | 0.127 | 0.0821 | No |
| 44 | Dffa | 6657 | 0.114 | 0.0685 | No |
| 45 | H1f0 | 6741 | 0.110 | 0.0675 | No |
| 46 | Cdk2 | 6778 | 0.110 | 0.0689 | No |
| 47 | Fdxr | 7084 | 0.097 | 0.0563 | No |
| 48 | Sod2 | 7138 | 0.094 | 0.0564 | No |
| 49 | Isg20 | 7443 | 0.081 | 0.0434 | No |
| 50 | Birc3 | 7646 | 0.072 | 0.0353 | No |
| 51 | Xiap | 7895 | 0.061 | 0.0246 | No |
| 52 | Bmf | 7942 | 0.059 | 0.0240 | No |
| 53 | Btg3 | 8122 | 0.052 | 0.0165 | No |
| 54 | Gpx1 | 8156 | 0.051 | 0.0163 | No |
| 55 | Wee1 | 8242 | 0.047 | 0.0134 | No |
| 56 | Aifm3 | 8276 | 0.045 | 0.0130 | No |
| 57 | Dap3 | 8410 | 0.040 | 0.0075 | No |
| 58 | Add1 | 8484 | 0.037 | 0.0049 | No |
| 59 | Pak1 | 8509 | 0.036 | 0.0047 | No |
| 60 | Atf3 | 8529 | 0.035 | 0.0047 | No |
| 61 | Ppt1 | 8779 | 0.023 | -0.0071 | No |
| 62 | Cflar | 8988 | 0.013 | -0.0172 | No |
| 63 | Gsr | 9190 | 0.004 | -0.0272 | No |
| 64 | Rock1 | 9373 | -0.002 | -0.0363 | No |
| 65 | Psen1 | 9413 | -0.004 | -0.0382 | No |
| 66 | Gpx4 | 9489 | -0.007 | -0.0417 | No |
| 67 | Cyld | 9628 | -0.014 | -0.0483 | No |
| 68 | Pea15a | 9736 | -0.019 | -0.0531 | No |
| 69 | Lmna | 9820 | -0.022 | -0.0566 | No |
| 70 | Smad7 | 9931 | -0.025 | -0.0615 | No |
| 71 | Dnm1l | 10053 | -0.031 | -0.0667 | No |
| 72 | Gsn | 10186 | -0.037 | -0.0722 | No |
| 73 | Crebbp | 10352 | -0.044 | -0.0793 | No |
| 74 | Tnfrsf12a | 10602 | -0.054 | -0.0902 | No |
| 75 | Cdc25b | 10686 | -0.057 | -0.0927 | No |
| 76 | Tap1 | 10753 | -0.060 | -0.0943 | No |
| 77 | Pmaip1 | 10901 | -0.067 | -0.0998 | No |
| 78 | Ppp2r5b | 11123 | -0.074 | -0.1088 | No |
| 79 | Dap | 11152 | -0.075 | -0.1080 | No |
| 80 | Tnf | 11302 | -0.081 | -0.1132 | No |
| 81 | Btg2 | 11381 | -0.085 | -0.1147 | No |
| 82 | Pdgfrb | 11401 | -0.086 | -0.1132 | No |
| 83 | Casp9 | 11475 | -0.089 | -0.1143 | No |
| 84 | Dpyd | 11589 | -0.093 | -0.1173 | No |
| 85 | Madd | 11606 | -0.094 | -0.1154 | No |
| 86 | Gadd45b | 11789 | -0.102 | -0.1216 | No |
| 87 | Casp4 | 11914 | -0.108 | -0.1247 | No |
| 88 | Vdac2 | 11919 | -0.108 | -0.1218 | No |
| 89 | Lgals3 | 12123 | -0.118 | -0.1286 | No |
| 90 | Plcb2 | 12214 | -0.120 | -0.1297 | No |
| 91 | Hgf | 12380 | -0.128 | -0.1343 | No |
| 92 | Bcl2l2 | 12409 | -0.129 | -0.1320 | No |
| 93 | Cd14 | 12437 | -0.130 | -0.1296 | No |
| 94 | Bcl2l11 | 12711 | -0.144 | -0.1392 | No |
| 95 | Tgfbr3 | 12800 | -0.148 | -0.1393 | No |
| 96 | Mmp2 | 12875 | -0.151 | -0.1387 | No |
| 97 | Dcn | 12946 | -0.155 | -0.1378 | No |
| 98 | Timp3 | 13187 | -0.165 | -0.1451 | No |
| 99 | Pdcd4 | 14270 | -0.216 | -0.1932 | No |
| 100 | Bax | 14403 | -0.224 | -0.1934 | No |
| 101 | Gpx3 | 14693 | -0.237 | -0.2011 | No |
| 102 | Nedd9 | 14716 | -0.239 | -0.1954 | No |
| 103 | Erbb3 | 14739 | -0.240 | -0.1896 | No |
| 104 | Gch1 | 14743 | -0.240 | -0.1828 | No |
| 105 | Casp6 | 14824 | -0.245 | -0.1798 | No |
| 106 | Bnip3l | 14829 | -0.245 | -0.1729 | No |
| 107 | Rela | 14836 | -0.245 | -0.1662 | No |
| 108 | Cd44 | 14858 | -0.246 | -0.1601 | No |
| 109 | Casp8 | 14939 | -0.250 | -0.1569 | No |
| 110 | Sc5d | 15014 | -0.253 | -0.1534 | No |
| 111 | Ccnd2 | 15171 | -0.260 | -0.1537 | No |
| 112 | Txnip | 15223 | -0.262 | -0.1488 | No |
| 113 | Bid | 15543 | -0.280 | -0.1567 | No |
| 114 | Psen2 | 15733 | -0.291 | -0.1579 | No |
| 115 | Cav1 | 15866 | -0.298 | -0.1559 | No |
| 116 | Ebp | 15945 | -0.303 | -0.1511 | No |
| 117 | Ifngr1 | 16526 | -0.334 | -0.1706 | No |
| 118 | Gstm2 | 17059 | -0.368 | -0.1868 | No |
| 119 | Casp7 | 17274 | -0.382 | -0.1865 | No |
| 120 | Eno2 | 17864 | -0.431 | -0.2037 | Yes |
| 121 | Satb1 | 17898 | -0.434 | -0.1929 | Yes |
| 122 | Tgfb2 | 18166 | -0.457 | -0.1932 | Yes |
| 123 | Casp3 | 18480 | -0.491 | -0.1948 | Yes |
| 124 | Bcl2l1 | 18615 | -0.506 | -0.1869 | Yes |
| 125 | Tnfsf10 | 18658 | -0.510 | -0.1744 | Yes |
| 126 | Mgmt | 18662 | -0.511 | -0.1598 | Yes |
| 127 | Ank | 18833 | -0.537 | -0.1529 | Yes |
| 128 | Ddit3 | 18849 | -0.539 | -0.1381 | Yes |
| 129 | Diablo | 19048 | -0.569 | -0.1317 | Yes |
| 130 | Fas | 19210 | -0.599 | -0.1226 | Yes |
| 131 | Timp2 | 19520 | -0.675 | -0.1187 | Yes |
| 132 | Krt18 | 19612 | -0.710 | -0.1028 | Yes |
| 133 | Il18 | 19697 | -0.743 | -0.0857 | Yes |
| 134 | Ier3 | 19736 | -0.767 | -0.0655 | Yes |
| 135 | Ptk2 | 19794 | -0.808 | -0.0451 | Yes |
| 136 | F2r | 19935 | -0.946 | -0.0249 | Yes |
| 137 | Ccnd1 | 19976 | -1.022 | 0.0025 | Yes |