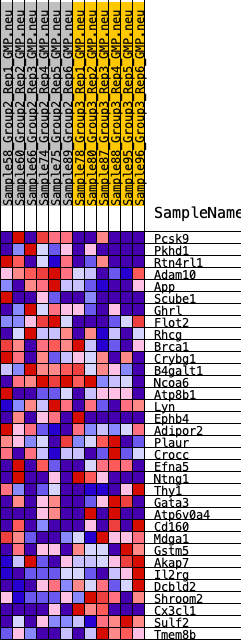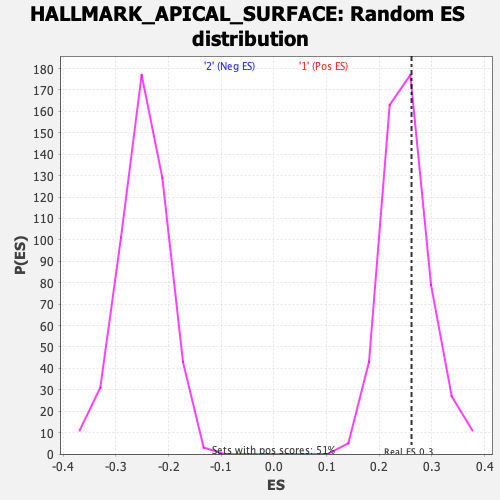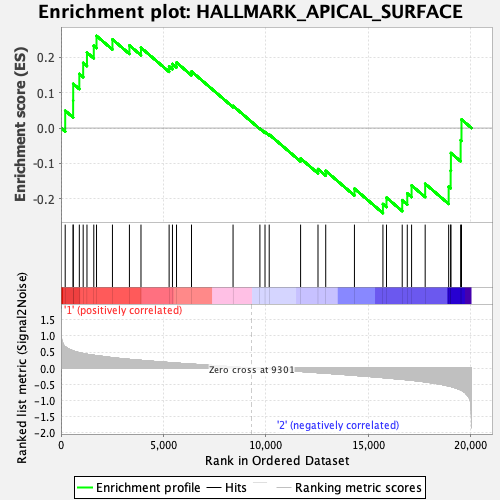
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | GMP.GMP.neu_Pheno.cls #Group2_versus_Group3.GMP.neu_Pheno.cls #Group2_versus_Group3_repos |
| Phenotype | GMP.neu_Pheno.cls#Group2_versus_Group3_repos |
| Upregulated in class | 1 |
| GeneSet | HALLMARK_APICAL_SURFACE |
| Enrichment Score (ES) | 0.2611982 |
| Normalized Enrichment Score (NES) | 1.0412449 |
| Nominal p-value | 0.37425742 |
| FDR q-value | 0.6387532 |
| FWER p-Value | 0.989 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Pcsk9 | 205 | 0.664 | 0.0492 | Yes |
| 2 | Pkhd1 | 592 | 0.535 | 0.0777 | Yes |
| 3 | Rtn4rl1 | 595 | 0.533 | 0.1254 | Yes |
| 4 | Adam10 | 894 | 0.480 | 0.1534 | Yes |
| 5 | App | 1082 | 0.454 | 0.1847 | Yes |
| 6 | Scube1 | 1267 | 0.431 | 0.2141 | Yes |
| 7 | Ghrl | 1600 | 0.399 | 0.2332 | Yes |
| 8 | Flot2 | 1733 | 0.387 | 0.2612 | Yes |
| 9 | Rhcg | 2510 | 0.323 | 0.2513 | No |
| 10 | Brca1 | 3338 | 0.272 | 0.2343 | No |
| 11 | Crybg1 | 3902 | 0.242 | 0.2278 | No |
| 12 | B4galt1 | 5275 | 0.174 | 0.1748 | No |
| 13 | Ncoa6 | 5438 | 0.166 | 0.1816 | No |
| 14 | Atp8b1 | 5636 | 0.157 | 0.1858 | No |
| 15 | Lyn | 6367 | 0.125 | 0.1604 | No |
| 16 | Ephb4 | 8394 | 0.040 | 0.0627 | No |
| 17 | Adipor2 | 9704 | -0.017 | -0.0012 | No |
| 18 | Plaur | 9953 | -0.026 | -0.0113 | No |
| 19 | Crocc | 10158 | -0.035 | -0.0183 | No |
| 20 | Efna5 | 11689 | -0.098 | -0.0861 | No |
| 21 | Ntng1 | 12538 | -0.137 | -0.1163 | No |
| 22 | Thy1 | 12915 | -0.154 | -0.1214 | No |
| 23 | Gata3 | 14316 | -0.220 | -0.1717 | No |
| 24 | Atp6v0a4 | 15706 | -0.289 | -0.2153 | No |
| 25 | Cd160 | 15880 | -0.299 | -0.1971 | No |
| 26 | Mdga1 | 16647 | -0.341 | -0.2049 | No |
| 27 | Gstm5 | 16894 | -0.357 | -0.1853 | No |
| 28 | Akap7 | 17104 | -0.370 | -0.1626 | No |
| 29 | Il2rg | 17767 | -0.423 | -0.1578 | No |
| 30 | Dcbld2 | 18913 | -0.547 | -0.1662 | No |
| 31 | Shroom2 | 19010 | -0.562 | -0.1207 | No |
| 32 | Cx3cl1 | 19023 | -0.564 | -0.0707 | No |
| 33 | Sulf2 | 19497 | -0.669 | -0.0345 | No |
| 34 | Tmem8b | 19539 | -0.680 | 0.0244 | No |