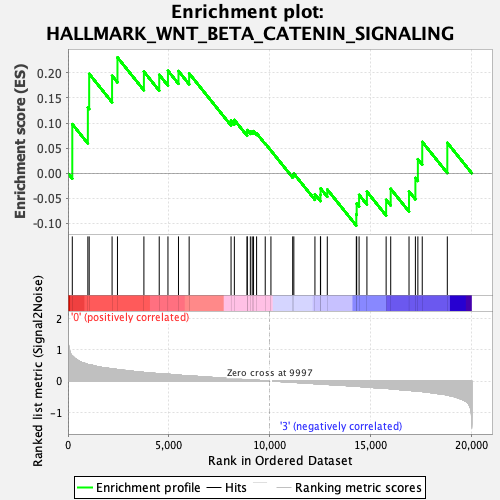
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | GMP.GMP.neu_Pheno.cls #Group1_versus_Group4.GMP.neu_Pheno.cls #Group1_versus_Group4_repos |
| Phenotype | GMP.neu_Pheno.cls#Group1_versus_Group4_repos |
| Upregulated in class | 0 |
| GeneSet | HALLMARK_WNT_BETA_CATENIN_SIGNALING |
| Enrichment Score (ES) | 0.23087437 |
| Normalized Enrichment Score (NES) | 0.8534505 |
| Nominal p-value | 0.6896552 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Tcf7 | 213 | 0.798 | 0.0980 | Yes |
| 2 | Jag2 | 985 | 0.526 | 0.1310 | Yes |
| 3 | Axin2 | 1050 | 0.517 | 0.1982 | Yes |
| 4 | Jag1 | 2186 | 0.389 | 0.1944 | Yes |
| 5 | Rbpj | 2453 | 0.366 | 0.2309 | Yes |
| 6 | Ppard | 3764 | 0.273 | 0.2026 | No |
| 7 | Wnt6 | 4524 | 0.233 | 0.1963 | No |
| 8 | Hdac5 | 4957 | 0.218 | 0.2043 | No |
| 9 | Cul1 | 5479 | 0.187 | 0.2037 | No |
| 10 | Ncor2 | 6008 | 0.160 | 0.1991 | No |
| 11 | Axin1 | 8086 | 0.074 | 0.1053 | No |
| 12 | Dvl2 | 8250 | 0.067 | 0.1063 | No |
| 13 | Ncstn | 8882 | 0.043 | 0.0805 | No |
| 14 | Ccnd2 | 8888 | 0.043 | 0.0861 | No |
| 15 | Notch4 | 9043 | 0.036 | 0.0833 | No |
| 16 | Adam17 | 9135 | 0.032 | 0.0831 | No |
| 17 | Nkd1 | 9201 | 0.030 | 0.0839 | No |
| 18 | Ctnnb1 | 9353 | 0.024 | 0.0796 | No |
| 19 | Notch1 | 9781 | 0.008 | 0.0594 | No |
| 20 | Gnai1 | 10064 | 0.000 | 0.0453 | No |
| 21 | Maml1 | 11143 | -0.038 | -0.0034 | No |
| 22 | Ptch1 | 11200 | -0.041 | -0.0007 | No |
| 23 | Hdac2 | 12246 | -0.082 | -0.0418 | No |
| 24 | Fzd1 | 12519 | -0.093 | -0.0427 | No |
| 25 | Hdac11 | 12527 | -0.094 | -0.0303 | No |
| 26 | Skp2 | 12859 | -0.107 | -0.0324 | No |
| 27 | Kat2a | 14298 | -0.164 | -0.0820 | No |
| 28 | Numb | 14317 | -0.165 | -0.0604 | No |
| 29 | Wnt5b | 14436 | -0.171 | -0.0431 | No |
| 30 | Csnk1e | 14824 | -0.189 | -0.0366 | No |
| 31 | Trp53 | 15777 | -0.230 | -0.0529 | No |
| 32 | Psen2 | 16004 | -0.244 | -0.0310 | No |
| 33 | Myc | 16912 | -0.295 | -0.0362 | No |
| 34 | Wnt1 | 17228 | -0.313 | -0.0093 | No |
| 35 | Lef1 | 17351 | -0.319 | 0.0281 | No |
| 36 | Frat1 | 17568 | -0.333 | 0.0626 | No |
| 37 | Fzd8 | 18811 | -0.443 | 0.0608 | No |