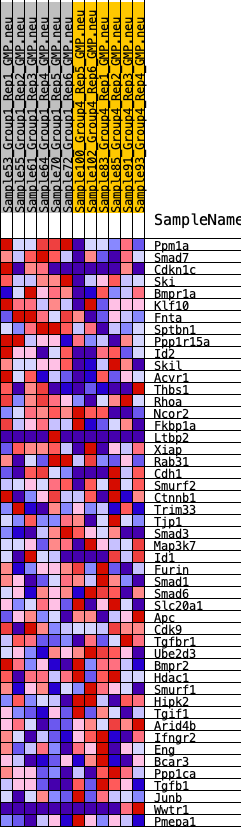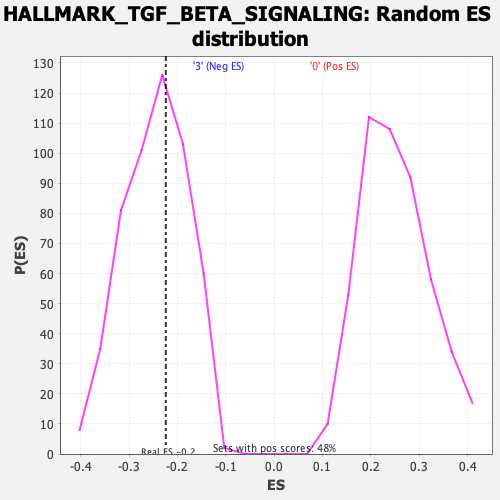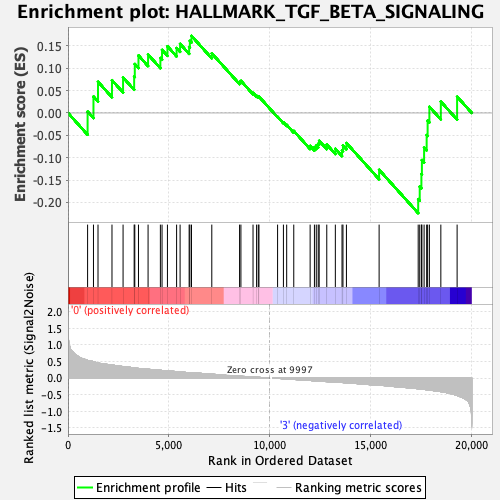
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | GMP.GMP.neu_Pheno.cls #Group1_versus_Group4.GMP.neu_Pheno.cls #Group1_versus_Group4_repos |
| Phenotype | GMP.neu_Pheno.cls#Group1_versus_Group4_repos |
| Upregulated in class | 3 |
| GeneSet | HALLMARK_TGF_BETA_SIGNALING |
| Enrichment Score (ES) | -0.22413976 |
| Normalized Enrichment Score (NES) | -0.914334 |
| Nominal p-value | 0.5872093 |
| FDR q-value | 0.82507926 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Ppm1a | 972 | 0.529 | 0.0031 | No |
| 2 | Smad7 | 1263 | 0.492 | 0.0367 | No |
| 3 | Cdkn1c | 1488 | 0.453 | 0.0698 | No |
| 4 | Ski | 2181 | 0.390 | 0.0733 | No |
| 5 | Bmpr1a | 2731 | 0.343 | 0.0794 | No |
| 6 | Klf10 | 3281 | 0.304 | 0.0816 | No |
| 7 | Fnta | 3313 | 0.302 | 0.1095 | No |
| 8 | Sptbn1 | 3495 | 0.288 | 0.1287 | No |
| 9 | Ppp1r15a | 3971 | 0.264 | 0.1308 | No |
| 10 | Id2 | 4581 | 0.233 | 0.1230 | No |
| 11 | Skil | 4664 | 0.227 | 0.1412 | No |
| 12 | Acvr1 | 4931 | 0.219 | 0.1493 | No |
| 13 | Thbs1 | 5386 | 0.192 | 0.1453 | No |
| 14 | Rhoa | 5559 | 0.183 | 0.1546 | No |
| 15 | Ncor2 | 6008 | 0.160 | 0.1478 | No |
| 16 | Fkbp1a | 6044 | 0.157 | 0.1615 | No |
| 17 | Ltbp2 | 6124 | 0.154 | 0.1726 | No |
| 18 | Xiap | 7131 | 0.115 | 0.1335 | No |
| 19 | Rab31 | 8509 | 0.056 | 0.0700 | No |
| 20 | Cdh1 | 8575 | 0.053 | 0.0719 | No |
| 21 | Smurf2 | 9176 | 0.031 | 0.0449 | No |
| 22 | Ctnnb1 | 9353 | 0.024 | 0.0384 | No |
| 23 | Trim33 | 9436 | 0.021 | 0.0364 | No |
| 24 | Tjp1 | 9470 | 0.020 | 0.0367 | No |
| 25 | Smad3 | 10394 | -0.009 | -0.0086 | No |
| 26 | Map3k7 | 10683 | -0.020 | -0.0210 | No |
| 27 | Id1 | 10845 | -0.025 | -0.0266 | No |
| 28 | Furin | 11196 | -0.040 | -0.0402 | No |
| 29 | Smad1 | 12008 | -0.072 | -0.0737 | No |
| 30 | Smad6 | 12221 | -0.081 | -0.0764 | No |
| 31 | Slc20a1 | 12312 | -0.084 | -0.0727 | No |
| 32 | Apc | 12408 | -0.089 | -0.0688 | No |
| 33 | Cdk9 | 12456 | -0.091 | -0.0622 | No |
| 34 | Tgfbr1 | 12833 | -0.106 | -0.0707 | No |
| 35 | Ube2d3 | 13258 | -0.121 | -0.0801 | No |
| 36 | Bmpr2 | 13589 | -0.133 | -0.0836 | No |
| 37 | Hdac1 | 13636 | -0.135 | -0.0727 | No |
| 38 | Smurf1 | 13808 | -0.143 | -0.0672 | No |
| 39 | Hipk2 | 15429 | -0.216 | -0.1272 | No |
| 40 | Tgif1 | 17367 | -0.321 | -0.1928 | Yes |
| 41 | Arid4b | 17443 | -0.326 | -0.1647 | Yes |
| 42 | Ifngr2 | 17526 | -0.330 | -0.1365 | Yes |
| 43 | Eng | 17551 | -0.332 | -0.1053 | Yes |
| 44 | Bcar3 | 17660 | -0.340 | -0.0775 | Yes |
| 45 | Ppp1ca | 17787 | -0.351 | -0.0494 | Yes |
| 46 | Tgfb1 | 17835 | -0.355 | -0.0171 | Yes |
| 47 | Junb | 17922 | -0.361 | 0.0139 | Yes |
| 48 | Wwtr1 | 18490 | -0.408 | 0.0255 | Yes |
| 49 | Pmepa1 | 19297 | -0.525 | 0.0365 | Yes |