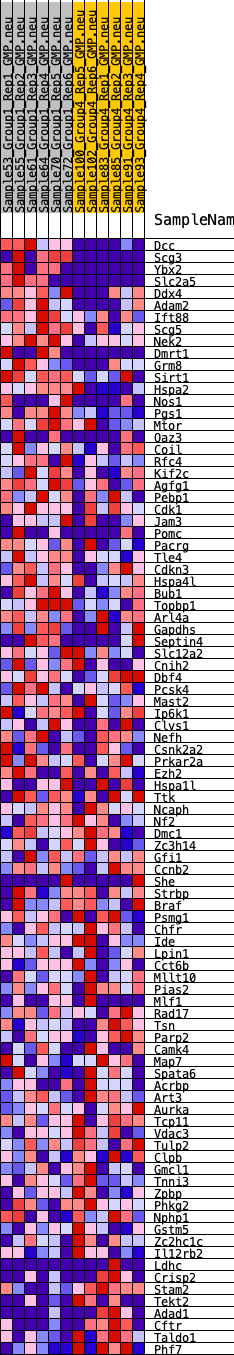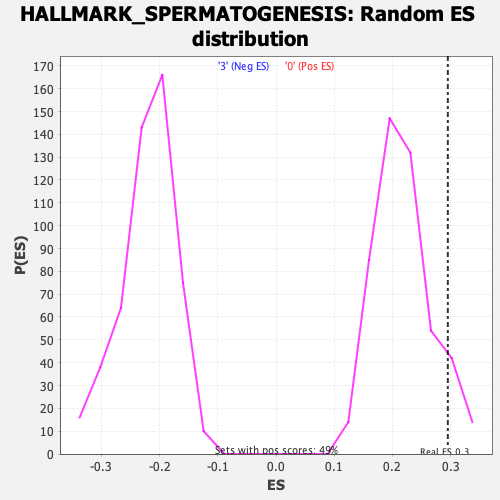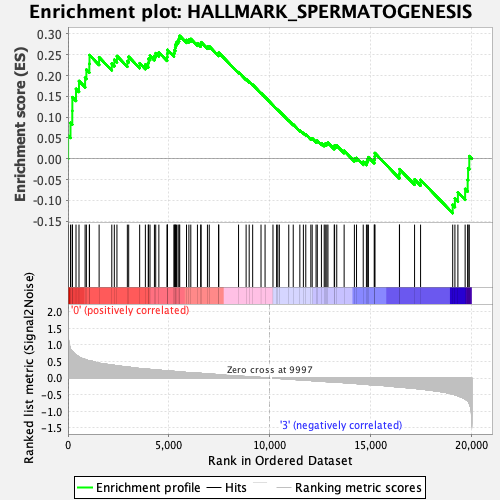
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | GMP.GMP.neu_Pheno.cls #Group1_versus_Group4.GMP.neu_Pheno.cls #Group1_versus_Group4_repos |
| Phenotype | GMP.neu_Pheno.cls#Group1_versus_Group4_repos |
| Upregulated in class | 0 |
| GeneSet | HALLMARK_SPERMATOGENESIS |
| Enrichment Score (ES) | 0.29535684 |
| Normalized Enrichment Score (NES) | 1.3582269 |
| Nominal p-value | 0.06967213 |
| FDR q-value | 0.83054733 |
| FWER p-Value | 0.66 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Dcc | 6 | 1.359 | 0.0560 | Yes |
| 2 | Scg3 | 124 | 0.875 | 0.0864 | Yes |
| 3 | Ybx2 | 212 | 0.799 | 0.1151 | Yes |
| 4 | Slc2a5 | 216 | 0.796 | 0.1480 | Yes |
| 5 | Ddx4 | 397 | 0.700 | 0.1679 | Yes |
| 6 | Adam2 | 545 | 0.635 | 0.1869 | Yes |
| 7 | Ift88 | 848 | 0.559 | 0.1949 | Yes |
| 8 | Scg5 | 911 | 0.543 | 0.2142 | Yes |
| 9 | Nek2 | 1058 | 0.516 | 0.2283 | Yes |
| 10 | Dmrt1 | 1067 | 0.514 | 0.2492 | Yes |
| 11 | Grm8 | 1543 | 0.447 | 0.2439 | Yes |
| 12 | Sirt1 | 2172 | 0.391 | 0.2286 | Yes |
| 13 | Hspa2 | 2297 | 0.381 | 0.2381 | Yes |
| 14 | Nos1 | 2427 | 0.369 | 0.2469 | Yes |
| 15 | Pgs1 | 2943 | 0.330 | 0.2348 | Yes |
| 16 | Mtor | 3010 | 0.327 | 0.2450 | Yes |
| 17 | Oaz3 | 3554 | 0.285 | 0.2296 | Yes |
| 18 | Coil | 3837 | 0.269 | 0.2266 | Yes |
| 19 | Rfc4 | 3979 | 0.263 | 0.2304 | Yes |
| 20 | Kif2c | 3997 | 0.263 | 0.2405 | Yes |
| 21 | Agfg1 | 4067 | 0.259 | 0.2477 | Yes |
| 22 | Pebp1 | 4286 | 0.245 | 0.2470 | Yes |
| 23 | Cdk1 | 4350 | 0.241 | 0.2538 | Yes |
| 24 | Jam3 | 4506 | 0.234 | 0.2557 | Yes |
| 25 | Pomc | 4913 | 0.220 | 0.2444 | Yes |
| 26 | Pacrg | 4927 | 0.219 | 0.2528 | Yes |
| 27 | Tle4 | 4934 | 0.219 | 0.2616 | Yes |
| 28 | Cdkn3 | 5252 | 0.199 | 0.2539 | Yes |
| 29 | Hspa4l | 5279 | 0.199 | 0.2609 | Yes |
| 30 | Bub1 | 5317 | 0.196 | 0.2671 | Yes |
| 31 | Topbp1 | 5331 | 0.195 | 0.2746 | Yes |
| 32 | Arl4a | 5382 | 0.193 | 0.2801 | Yes |
| 33 | Gapdhs | 5456 | 0.188 | 0.2842 | Yes |
| 34 | Septin4 | 5507 | 0.186 | 0.2894 | Yes |
| 35 | Slc12a2 | 5541 | 0.184 | 0.2954 | Yes |
| 36 | Cnih2 | 5877 | 0.166 | 0.2854 | No |
| 37 | Dbf4 | 5995 | 0.161 | 0.2862 | No |
| 38 | Pcsk4 | 6090 | 0.155 | 0.2879 | No |
| 39 | Mast2 | 6419 | 0.146 | 0.2776 | No |
| 40 | Ip6k1 | 6580 | 0.140 | 0.2753 | No |
| 41 | Clvs1 | 6601 | 0.139 | 0.2801 | No |
| 42 | Nefh | 6915 | 0.125 | 0.2696 | No |
| 43 | Csnk2a2 | 7005 | 0.121 | 0.2701 | No |
| 44 | Prkar2a | 7466 | 0.101 | 0.2512 | No |
| 45 | Ezh2 | 7478 | 0.101 | 0.2548 | No |
| 46 | Hspa1l | 8458 | 0.059 | 0.2081 | No |
| 47 | Ttk | 8833 | 0.044 | 0.1912 | No |
| 48 | Ncaph | 8985 | 0.039 | 0.1852 | No |
| 49 | Nf2 | 9158 | 0.031 | 0.1779 | No |
| 50 | Dmc1 | 9572 | 0.016 | 0.1579 | No |
| 51 | Zc3h14 | 9773 | 0.008 | 0.1482 | No |
| 52 | Gfi1 | 10165 | -0.000 | 0.1286 | No |
| 53 | Ccnb2 | 10345 | -0.007 | 0.1199 | No |
| 54 | She | 10404 | -0.010 | 0.1174 | No |
| 55 | Strbp | 10481 | -0.012 | 0.1141 | No |
| 56 | Braf | 10948 | -0.030 | 0.0919 | No |
| 57 | Psmg1 | 11170 | -0.039 | 0.0825 | No |
| 58 | Chfr | 11498 | -0.052 | 0.0682 | No |
| 59 | Ide | 11673 | -0.059 | 0.0619 | No |
| 60 | Lpin1 | 11792 | -0.064 | 0.0587 | No |
| 61 | Cct6b | 12043 | -0.074 | 0.0492 | No |
| 62 | Mllt10 | 12109 | -0.077 | 0.0491 | No |
| 63 | Pias2 | 12302 | -0.083 | 0.0429 | No |
| 64 | Mlf1 | 12363 | -0.086 | 0.0435 | No |
| 65 | Rad17 | 12568 | -0.095 | 0.0372 | No |
| 66 | Tsn | 12706 | -0.102 | 0.0345 | No |
| 67 | Parp2 | 12758 | -0.104 | 0.0363 | No |
| 68 | Camk4 | 12834 | -0.106 | 0.0369 | No |
| 69 | Map7 | 12891 | -0.108 | 0.0386 | No |
| 70 | Spata6 | 13203 | -0.119 | 0.0279 | No |
| 71 | Acrbp | 13221 | -0.119 | 0.0320 | No |
| 72 | Art3 | 13328 | -0.123 | 0.0318 | No |
| 73 | Aurka | 13694 | -0.138 | 0.0192 | No |
| 74 | Tcp11 | 14203 | -0.160 | 0.0003 | No |
| 75 | Vdac3 | 14309 | -0.164 | 0.0018 | No |
| 76 | Tulp2 | 14641 | -0.180 | -0.0073 | No |
| 77 | Clpb | 14799 | -0.188 | -0.0074 | No |
| 78 | Gmcl1 | 14844 | -0.191 | -0.0017 | No |
| 79 | Tnni3 | 14898 | -0.194 | 0.0037 | No |
| 80 | Zpbp | 15186 | -0.208 | -0.0021 | No |
| 81 | Phkg2 | 15211 | -0.208 | 0.0053 | No |
| 82 | Nphp1 | 15218 | -0.209 | 0.0137 | No |
| 83 | Gstm5 | 16436 | -0.270 | -0.0362 | No |
| 84 | Zc2hc1c | 16441 | -0.270 | -0.0252 | No |
| 85 | Il12rb2 | 17187 | -0.311 | -0.0496 | No |
| 86 | Ldhc | 17485 | -0.328 | -0.0509 | No |
| 87 | Crisp2 | 19078 | -0.481 | -0.1109 | No |
| 88 | Stam2 | 19188 | -0.503 | -0.0955 | No |
| 89 | Tekt2 | 19336 | -0.533 | -0.0808 | No |
| 90 | Adad1 | 19695 | -0.631 | -0.0726 | No |
| 91 | Cftr | 19820 | -0.684 | -0.0505 | No |
| 92 | Taldo1 | 19842 | -0.698 | -0.0226 | No |
| 93 | Phf7 | 19901 | -0.768 | 0.0063 | No |