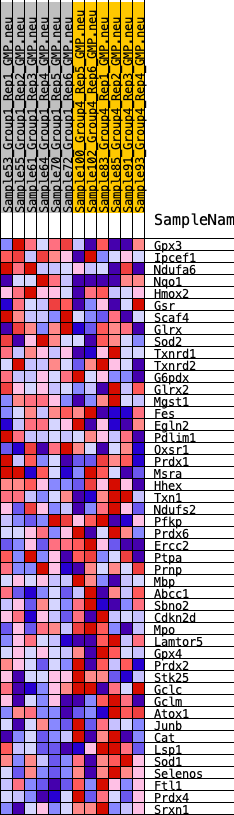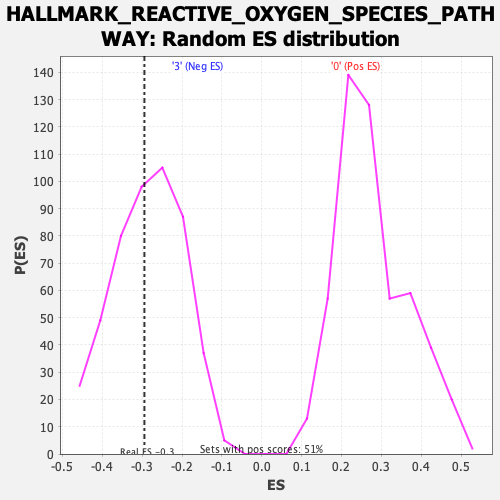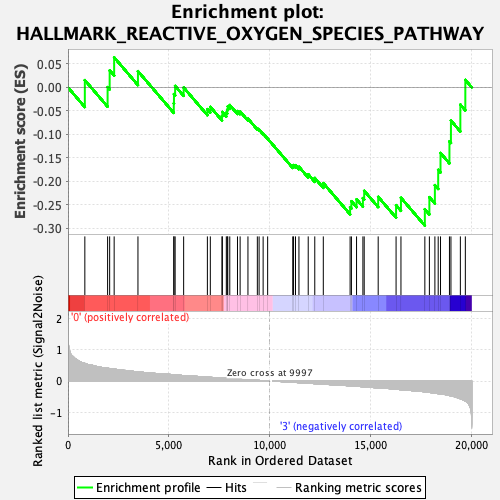
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | GMP.GMP.neu_Pheno.cls #Group1_versus_Group4.GMP.neu_Pheno.cls #Group1_versus_Group4_repos |
| Phenotype | GMP.neu_Pheno.cls#Group1_versus_Group4_repos |
| Upregulated in class | 3 |
| GeneSet | HALLMARK_REACTIVE_OXYGEN_SPECIES_PATHWAY |
| Enrichment Score (ES) | -0.29431129 |
| Normalized Enrichment Score (NES) | -1.035832 |
| Nominal p-value | 0.436214 |
| FDR q-value | 0.7079817 |
| FWER p-Value | 0.998 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Gpx3 | 835 | 0.562 | 0.0156 | No |
| 2 | Ipcef1 | 1964 | 0.407 | 0.0006 | No |
| 3 | Ndufa6 | 2066 | 0.398 | 0.0362 | No |
| 4 | Nqo1 | 2289 | 0.381 | 0.0640 | No |
| 5 | Hmox2 | 3469 | 0.291 | 0.0347 | No |
| 6 | Gsr | 5246 | 0.199 | -0.0339 | No |
| 7 | Scaf4 | 5256 | 0.199 | -0.0140 | No |
| 8 | Glrx | 5313 | 0.197 | 0.0032 | No |
| 9 | Sod2 | 5735 | 0.174 | -0.0001 | No |
| 10 | Txnrd1 | 6912 | 0.125 | -0.0463 | No |
| 11 | Txnrd2 | 7058 | 0.118 | -0.0415 | No |
| 12 | G6pdx | 7636 | 0.094 | -0.0608 | No |
| 13 | Glrx2 | 7655 | 0.093 | -0.0522 | No |
| 14 | Mgst1 | 7851 | 0.083 | -0.0535 | No |
| 15 | Fes | 7897 | 0.082 | -0.0474 | No |
| 16 | Egln2 | 7923 | 0.081 | -0.0404 | No |
| 17 | Pdlim1 | 8023 | 0.077 | -0.0375 | No |
| 18 | Oxsr1 | 8405 | 0.061 | -0.0503 | No |
| 19 | Prdx1 | 8536 | 0.054 | -0.0513 | No |
| 20 | Msra | 8924 | 0.041 | -0.0665 | No |
| 21 | Hhex | 9390 | 0.023 | -0.0874 | No |
| 22 | Txn1 | 9477 | 0.020 | -0.0897 | No |
| 23 | Ndufs2 | 9677 | 0.012 | -0.0984 | No |
| 24 | Pfkp | 9901 | 0.004 | -0.1092 | No |
| 25 | Prdx6 | 11146 | -0.038 | -0.1676 | No |
| 26 | Ercc2 | 11186 | -0.040 | -0.1654 | No |
| 27 | Ptpa | 11282 | -0.044 | -0.1657 | No |
| 28 | Prnp | 11453 | -0.050 | -0.1691 | No |
| 29 | Mbp | 11913 | -0.069 | -0.1850 | No |
| 30 | Abcc1 | 12238 | -0.082 | -0.1929 | No |
| 31 | Sbno2 | 12661 | -0.100 | -0.2038 | No |
| 32 | Cdkn2d | 13990 | -0.151 | -0.2548 | No |
| 33 | Mpo | 14056 | -0.154 | -0.2424 | No |
| 34 | Lamtor5 | 14310 | -0.164 | -0.2383 | No |
| 35 | Gpx4 | 14625 | -0.180 | -0.2357 | No |
| 36 | Prdx2 | 14689 | -0.183 | -0.2202 | No |
| 37 | Stk25 | 15383 | -0.214 | -0.2331 | No |
| 38 | Gclc | 16270 | -0.261 | -0.2508 | No |
| 39 | Gclm | 16512 | -0.273 | -0.2350 | No |
| 40 | Atox1 | 17697 | -0.342 | -0.2594 | Yes |
| 41 | Junb | 17922 | -0.361 | -0.2338 | Yes |
| 42 | Cat | 18195 | -0.385 | -0.2081 | Yes |
| 43 | Lsp1 | 18359 | -0.399 | -0.1755 | Yes |
| 44 | Sod1 | 18474 | -0.408 | -0.1397 | Yes |
| 45 | Selenos | 18917 | -0.460 | -0.1148 | Yes |
| 46 | Ftl1 | 18991 | -0.469 | -0.0706 | Yes |
| 47 | Prdx4 | 19458 | -0.562 | -0.0365 | Yes |
| 48 | Srxn1 | 19705 | -0.636 | 0.0161 | Yes |