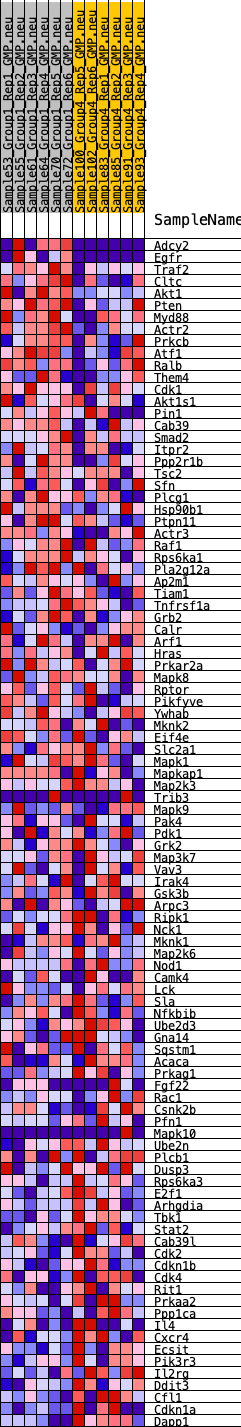
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | GMP.GMP.neu_Pheno.cls #Group1_versus_Group4.GMP.neu_Pheno.cls #Group1_versus_Group4_repos |
| Phenotype | GMP.neu_Pheno.cls#Group1_versus_Group4_repos |
| Upregulated in class | 3 |
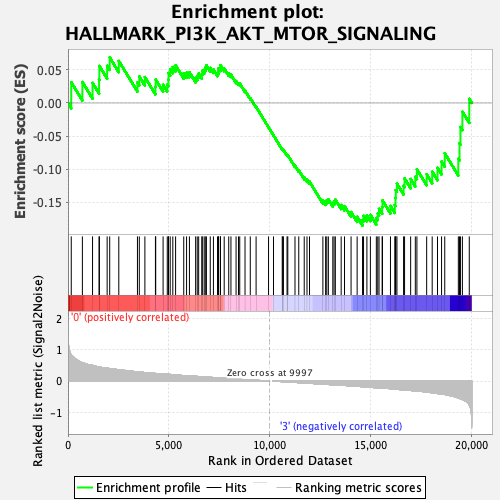
| GeneSet | HALLMARK_PI3K_AKT_MTOR_SIGNALING |
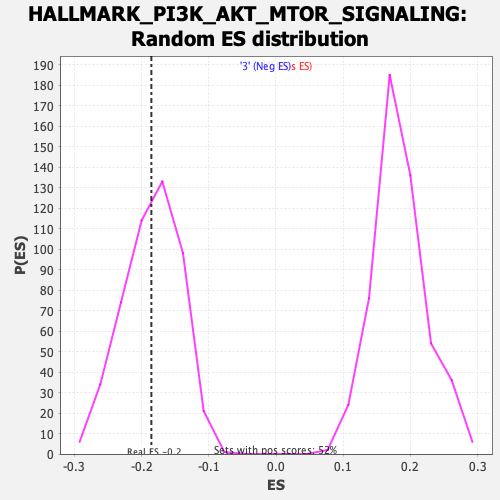
| Enrichment Score (ES) | -0.18535395 |
| Normalized Enrichment Score (NES) | -1.0047024 |
| Nominal p-value | 0.46361747 |
| FDR q-value | 0.706786 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Adcy2 | 161 | 0.829 | 0.0314 | No |
| 2 | Egfr | 713 | 0.588 | 0.0318 | No |
| 3 | Traf2 | 1217 | 0.500 | 0.0304 | No |
| 4 | Cltc | 1541 | 0.447 | 0.0355 | No |
| 5 | Akt1 | 1556 | 0.445 | 0.0560 | No |
| 6 | Pten | 1941 | 0.410 | 0.0563 | No |
| 7 | Myd88 | 2068 | 0.398 | 0.0689 | No |
| 8 | Actr2 | 2519 | 0.362 | 0.0636 | No |
| 9 | Prkcb | 3444 | 0.292 | 0.0312 | No |
| 10 | Atf1 | 3536 | 0.286 | 0.0402 | No |
| 11 | Ralb | 3813 | 0.271 | 0.0393 | No |
| 12 | Them4 | 4336 | 0.242 | 0.0246 | No |
| 13 | Cdk1 | 4350 | 0.241 | 0.0354 | No |
| 14 | Akt1s1 | 4716 | 0.224 | 0.0277 | No |
| 15 | Pin1 | 4923 | 0.220 | 0.0279 | No |
| 16 | Cab39 | 4980 | 0.216 | 0.0353 | No |
| 17 | Smad2 | 4985 | 0.215 | 0.0454 | No |
| 18 | Itpr2 | 5068 | 0.211 | 0.0513 | No |
| 19 | Ppp2r1b | 5198 | 0.203 | 0.0545 | No |
| 20 | Tsc2 | 5336 | 0.195 | 0.0569 | No |
| 21 | Sfn | 5740 | 0.173 | 0.0450 | No |
| 22 | Plcg1 | 5880 | 0.166 | 0.0459 | No |
| 23 | Hsp90b1 | 6022 | 0.159 | 0.0464 | No |
| 24 | Ptpn11 | 6330 | 0.150 | 0.0381 | No |
| 25 | Actr3 | 6409 | 0.147 | 0.0412 | No |
| 26 | Raf1 | 6480 | 0.145 | 0.0446 | No |
| 27 | Rps6ka1 | 6637 | 0.137 | 0.0433 | No |
| 28 | Pla2g12a | 6659 | 0.136 | 0.0488 | No |
| 29 | Ap2m1 | 6752 | 0.131 | 0.0504 | No |
| 30 | Tiam1 | 6808 | 0.128 | 0.0537 | No |
| 31 | Tnfrsf1a | 6864 | 0.127 | 0.0570 | No |
| 32 | Grb2 | 7055 | 0.118 | 0.0531 | No |
| 33 | Calr | 7212 | 0.112 | 0.0506 | No |
| 34 | Arf1 | 7415 | 0.103 | 0.0454 | No |
| 35 | Hras | 7461 | 0.101 | 0.0480 | No |
| 36 | Prkar2a | 7466 | 0.101 | 0.0526 | No |
| 37 | Mapk8 | 7556 | 0.098 | 0.0528 | No |
| 38 | Rptor | 7561 | 0.098 | 0.0572 | No |
| 39 | Pikfyve | 7746 | 0.088 | 0.0522 | No |
| 40 | Ywhab | 7971 | 0.079 | 0.0447 | No |
| 41 | Mknk2 | 8081 | 0.075 | 0.0428 | No |
| 42 | Eif4e | 8335 | 0.064 | 0.0332 | No |
| 43 | Slc2a1 | 8455 | 0.059 | 0.0300 | No |
| 44 | Mapk1 | 8517 | 0.056 | 0.0296 | No |
| 45 | Mapkap1 | 8779 | 0.046 | 0.0187 | No |
| 46 | Map2k3 | 9039 | 0.036 | 0.0074 | No |
| 47 | Trib3 | 9328 | 0.024 | -0.0059 | No |
| 48 | Mapk9 | 9946 | 0.002 | -0.0367 | No |
| 49 | Pak4 | 10191 | -0.001 | -0.0489 | No |
| 50 | Pdk1 | 10624 | -0.018 | -0.0697 | No |
| 51 | Grk2 | 10647 | -0.019 | -0.0699 | No |
| 52 | Map3k7 | 10683 | -0.020 | -0.0707 | No |
| 53 | Vav3 | 10863 | -0.026 | -0.0785 | No |
| 54 | Irak4 | 10900 | -0.027 | -0.0790 | No |
| 55 | Gsk3b | 11257 | -0.043 | -0.0948 | No |
| 56 | Arpc3 | 11444 | -0.050 | -0.1017 | No |
| 57 | Ripk1 | 11715 | -0.061 | -0.1124 | No |
| 58 | Nck1 | 11852 | -0.067 | -0.1160 | No |
| 59 | Mknk1 | 11977 | -0.071 | -0.1189 | No |
| 60 | Map2k6 | 12640 | -0.099 | -0.1474 | No |
| 61 | Nod1 | 12776 | -0.104 | -0.1492 | No |
| 62 | Camk4 | 12834 | -0.106 | -0.1470 | No |
| 63 | Lck | 12912 | -0.109 | -0.1457 | No |
| 64 | Sla | 13133 | -0.116 | -0.1512 | No |
| 65 | Nfkbib | 13194 | -0.118 | -0.1486 | No |
| 66 | Ube2d3 | 13258 | -0.121 | -0.1460 | No |
| 67 | Gna14 | 13547 | -0.131 | -0.1542 | No |
| 68 | Sqstm1 | 13713 | -0.138 | -0.1559 | No |
| 69 | Acaca | 14038 | -0.153 | -0.1648 | No |
| 70 | Prkag1 | 14339 | -0.166 | -0.1720 | No |
| 71 | Fgf22 | 14607 | -0.179 | -0.1768 | Yes |
| 72 | Rac1 | 14656 | -0.181 | -0.1706 | Yes |
| 73 | Csnk2b | 14823 | -0.189 | -0.1699 | Yes |
| 74 | Pfn1 | 14996 | -0.199 | -0.1691 | Yes |
| 75 | Mapk10 | 15291 | -0.211 | -0.1738 | Yes |
| 76 | Ube2n | 15362 | -0.212 | -0.1672 | Yes |
| 77 | Plcb1 | 15424 | -0.216 | -0.1599 | Yes |
| 78 | Dusp3 | 15579 | -0.222 | -0.1571 | Yes |
| 79 | Rps6ka3 | 15596 | -0.222 | -0.1473 | Yes |
| 80 | E2f1 | 15992 | -0.243 | -0.1555 | Yes |
| 81 | Arhgdia | 16207 | -0.257 | -0.1540 | Yes |
| 82 | Tbk1 | 16241 | -0.259 | -0.1433 | Yes |
| 83 | Stat2 | 16256 | -0.260 | -0.1317 | Yes |
| 84 | Cab39l | 16313 | -0.264 | -0.1219 | Yes |
| 85 | Cdk2 | 16642 | -0.279 | -0.1251 | Yes |
| 86 | Cdkn1b | 16691 | -0.283 | -0.1140 | Yes |
| 87 | Cdk4 | 16993 | -0.301 | -0.1148 | Yes |
| 88 | Rit1 | 17224 | -0.313 | -0.1114 | Yes |
| 89 | Prkaa2 | 17303 | -0.317 | -0.1002 | Yes |
| 90 | Ppp1ca | 17787 | -0.351 | -0.1077 | Yes |
| 91 | Il4 | 18061 | -0.372 | -0.1037 | Yes |
| 92 | Cxcr4 | 18324 | -0.396 | -0.0980 | Yes |
| 93 | Ecsit | 18526 | -0.412 | -0.0884 | Yes |
| 94 | Pik3r3 | 18684 | -0.427 | -0.0760 | Yes |
| 95 | Il2rg | 19358 | -0.537 | -0.0841 | Yes |
| 96 | Ddit3 | 19416 | -0.550 | -0.0608 | Yes |
| 97 | Cfl1 | 19462 | -0.563 | -0.0362 | Yes |
| 98 | Cdkn1a | 19559 | -0.586 | -0.0131 | Yes |
| 99 | Dapp1 | 19900 | -0.766 | 0.0063 | Yes |