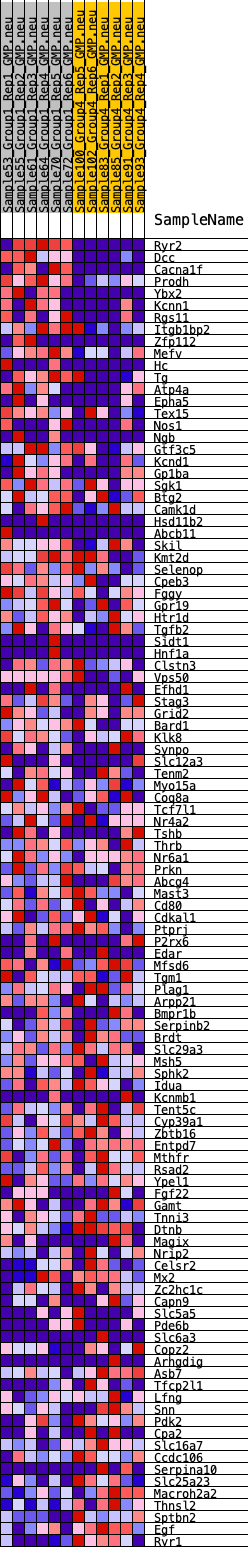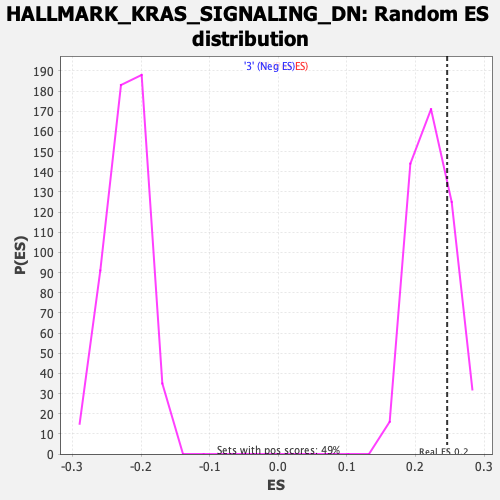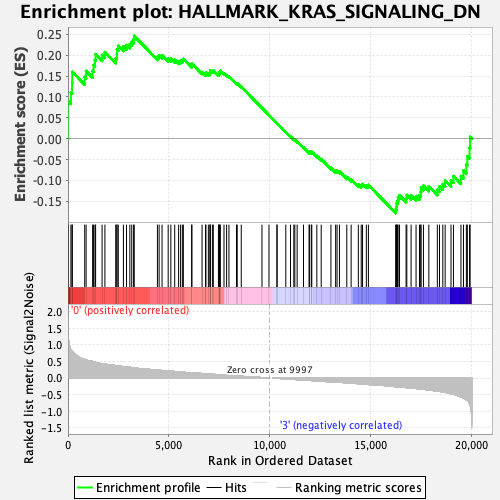
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | GMP.GMP.neu_Pheno.cls #Group1_versus_Group4.GMP.neu_Pheno.cls #Group1_versus_Group4_repos |
| Phenotype | GMP.neu_Pheno.cls#Group1_versus_Group4_repos |
| Upregulated in class | 0 |
| GeneSet | HALLMARK_KRAS_SIGNALING_DN |
| Enrichment Score (ES) | 0.24617974 |
| Normalized Enrichment Score (NES) | 1.1028798 |
| Nominal p-value | 0.20696722 |
| FDR q-value | 0.88401645 |
| FWER p-Value | 0.965 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Ryr2 | 5 | 1.390 | 0.0454 | Yes |
| 2 | Dcc | 6 | 1.359 | 0.0899 | Yes |
| 3 | Cacna1f | 144 | 0.849 | 0.1109 | Yes |
| 4 | Prodh | 203 | 0.805 | 0.1344 | Yes |
| 5 | Ybx2 | 212 | 0.799 | 0.1602 | Yes |
| 6 | Kcnn1 | 829 | 0.563 | 0.1478 | Yes |
| 7 | Rgs11 | 898 | 0.545 | 0.1622 | Yes |
| 8 | Itgb1bp2 | 1221 | 0.499 | 0.1624 | Yes |
| 9 | Zfp112 | 1268 | 0.491 | 0.1763 | Yes |
| 10 | Mefv | 1337 | 0.476 | 0.1885 | Yes |
| 11 | Hc | 1369 | 0.470 | 0.2023 | Yes |
| 12 | Tg | 1690 | 0.431 | 0.2004 | Yes |
| 13 | Atp4a | 1830 | 0.421 | 0.2072 | Yes |
| 14 | Epha5 | 2367 | 0.373 | 0.1926 | Yes |
| 15 | Tex15 | 2423 | 0.369 | 0.2019 | Yes |
| 16 | Nos1 | 2427 | 0.369 | 0.2138 | Yes |
| 17 | Ngb | 2494 | 0.362 | 0.2224 | Yes |
| 18 | Gtf3c5 | 2748 | 0.341 | 0.2209 | Yes |
| 19 | Kcnd1 | 2903 | 0.334 | 0.2242 | Yes |
| 20 | Gp1ba | 3067 | 0.322 | 0.2265 | Yes |
| 21 | Sgk1 | 3161 | 0.314 | 0.2322 | Yes |
| 22 | Btg2 | 3254 | 0.306 | 0.2376 | Yes |
| 23 | Camk1d | 3283 | 0.304 | 0.2462 | Yes |
| 24 | Hsd11b2 | 4435 | 0.235 | 0.1961 | No |
| 25 | Abcb11 | 4519 | 0.233 | 0.1996 | No |
| 26 | Skil | 4664 | 0.227 | 0.1998 | No |
| 27 | Kmt2d | 4968 | 0.217 | 0.1917 | No |
| 28 | Selenop | 5100 | 0.208 | 0.1920 | No |
| 29 | Cpeb3 | 5292 | 0.198 | 0.1889 | No |
| 30 | Fggy | 5486 | 0.187 | 0.1853 | No |
| 31 | Gpr19 | 5583 | 0.182 | 0.1865 | No |
| 32 | Htr1d | 5671 | 0.177 | 0.1879 | No |
| 33 | Tgfb2 | 5721 | 0.174 | 0.1911 | No |
| 34 | Sidt1 | 6133 | 0.154 | 0.1756 | No |
| 35 | Hnf1a | 6144 | 0.154 | 0.1801 | No |
| 36 | Clstn3 | 6648 | 0.137 | 0.1593 | No |
| 37 | Vps50 | 6821 | 0.127 | 0.1549 | No |
| 38 | Efhd1 | 6839 | 0.127 | 0.1582 | No |
| 39 | Stag3 | 6961 | 0.123 | 0.1562 | No |
| 40 | Grid2 | 7043 | 0.119 | 0.1560 | No |
| 41 | Bard1 | 7053 | 0.118 | 0.1594 | No |
| 42 | Klk8 | 7054 | 0.118 | 0.1633 | No |
| 43 | Synpo | 7161 | 0.114 | 0.1617 | No |
| 44 | Slc12a3 | 7205 | 0.112 | 0.1633 | No |
| 45 | Tenm2 | 7460 | 0.101 | 0.1538 | No |
| 46 | Myo15a | 7495 | 0.100 | 0.1554 | No |
| 47 | Coq8a | 7504 | 0.100 | 0.1583 | No |
| 48 | Tcf7l1 | 7523 | 0.099 | 0.1606 | No |
| 49 | Nr4a2 | 7555 | 0.098 | 0.1623 | No |
| 50 | Tshb | 7736 | 0.089 | 0.1561 | No |
| 51 | Thrb | 7866 | 0.083 | 0.1524 | No |
| 52 | Nr6a1 | 7982 | 0.078 | 0.1492 | No |
| 53 | Prkn | 8356 | 0.063 | 0.1325 | No |
| 54 | Abcg4 | 8392 | 0.062 | 0.1328 | No |
| 55 | Mast3 | 8592 | 0.052 | 0.1245 | No |
| 56 | Cd80 | 9620 | 0.014 | 0.0734 | No |
| 57 | Cdkal1 | 9967 | 0.001 | 0.0561 | No |
| 58 | Ptprj | 10362 | -0.008 | 0.0366 | No |
| 59 | P2rx6 | 10368 | -0.008 | 0.0366 | No |
| 60 | Edar | 10801 | -0.024 | 0.0157 | No |
| 61 | Mfsd6 | 11035 | -0.034 | 0.0051 | No |
| 62 | Tgm1 | 11199 | -0.041 | -0.0018 | No |
| 63 | Plag1 | 11249 | -0.043 | -0.0028 | No |
| 64 | Arpp21 | 11364 | -0.047 | -0.0070 | No |
| 65 | Bmpr1b | 11679 | -0.059 | -0.0208 | No |
| 66 | Serpinb2 | 11962 | -0.070 | -0.0327 | No |
| 67 | Brdt | 11966 | -0.070 | -0.0305 | No |
| 68 | Slc29a3 | 12064 | -0.075 | -0.0330 | No |
| 69 | Msh5 | 12090 | -0.076 | -0.0317 | No |
| 70 | Sphk2 | 12340 | -0.085 | -0.0414 | No |
| 71 | Idua | 12557 | -0.095 | -0.0492 | No |
| 72 | Kcnmb1 | 13040 | -0.114 | -0.0696 | No |
| 73 | Tent5c | 13273 | -0.121 | -0.0773 | No |
| 74 | Cyp39a1 | 13343 | -0.124 | -0.0767 | No |
| 75 | Zbtb16 | 13462 | -0.128 | -0.0784 | No |
| 76 | Entpd7 | 13826 | -0.144 | -0.0919 | No |
| 77 | Mthfr | 14041 | -0.153 | -0.0976 | No |
| 78 | Rsad2 | 14398 | -0.169 | -0.1099 | No |
| 79 | Ypel1 | 14541 | -0.176 | -0.1113 | No |
| 80 | Fgf22 | 14607 | -0.179 | -0.1087 | No |
| 81 | Gamt | 14796 | -0.188 | -0.1120 | No |
| 82 | Tnni3 | 14898 | -0.194 | -0.1107 | No |
| 83 | Dtnb | 16249 | -0.260 | -0.1699 | No |
| 84 | Magix | 16290 | -0.262 | -0.1633 | No |
| 85 | Nrip2 | 16293 | -0.262 | -0.1548 | No |
| 86 | Celsr2 | 16343 | -0.265 | -0.1486 | No |
| 87 | Mx2 | 16369 | -0.266 | -0.1411 | No |
| 88 | Zc2hc1c | 16441 | -0.270 | -0.1358 | No |
| 89 | Capn9 | 16767 | -0.287 | -0.1427 | No |
| 90 | Slc5a5 | 16803 | -0.289 | -0.1350 | No |
| 91 | Pde6b | 17014 | -0.301 | -0.1356 | No |
| 92 | Slc6a3 | 17261 | -0.316 | -0.1376 | No |
| 93 | Copz2 | 17432 | -0.325 | -0.1355 | No |
| 94 | Arhgdig | 17495 | -0.328 | -0.1278 | No |
| 95 | Asb7 | 17507 | -0.328 | -0.1176 | No |
| 96 | Tfcp2l1 | 17627 | -0.337 | -0.1125 | No |
| 97 | Lfng | 17895 | -0.358 | -0.1142 | No |
| 98 | Snn | 18316 | -0.396 | -0.1223 | No |
| 99 | Pdk2 | 18424 | -0.404 | -0.1144 | No |
| 100 | Cpa2 | 18588 | -0.419 | -0.1088 | No |
| 101 | Slc16a7 | 18704 | -0.430 | -0.1005 | No |
| 102 | Ccdc106 | 18994 | -0.470 | -0.0996 | No |
| 103 | Serpina10 | 19115 | -0.488 | -0.0896 | No |
| 104 | Slc25a23 | 19488 | -0.568 | -0.0896 | No |
| 105 | Macroh2a2 | 19612 | -0.599 | -0.0761 | No |
| 106 | Thnsl2 | 19756 | -0.653 | -0.0619 | No |
| 107 | Sptbn2 | 19804 | -0.675 | -0.0421 | No |
| 108 | Egf | 19914 | -0.782 | -0.0219 | No |
| 109 | Ryr1 | 19941 | -0.837 | 0.0043 | No |