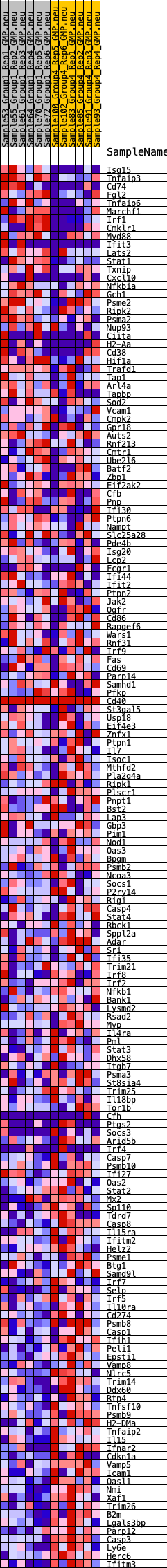
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | GMP.GMP.neu_Pheno.cls #Group1_versus_Group4.GMP.neu_Pheno.cls #Group1_versus_Group4_repos |
| Phenotype | GMP.neu_Pheno.cls#Group1_versus_Group4_repos |
| Upregulated in class | 3 |
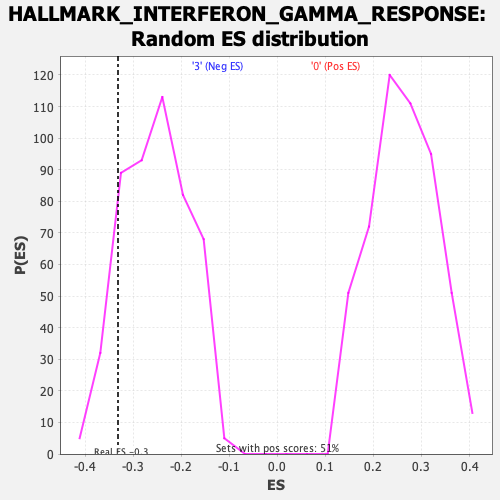
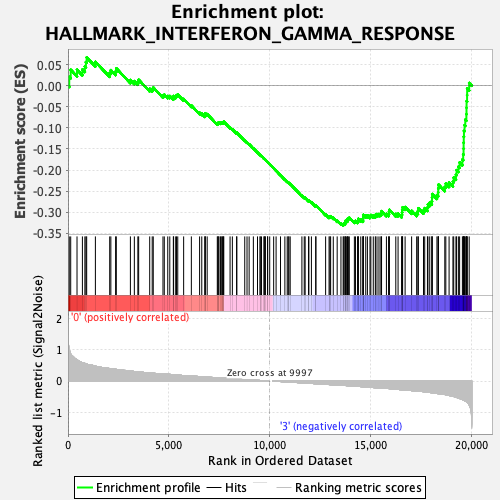
| GeneSet | HALLMARK_INTERFERON_GAMMA_RESPONSE |
| Enrichment Score (ES) | -0.33182493 |
| Normalized Enrichment Score (NES) | -1.3039092 |
| Nominal p-value | 0.13347022 |
| FDR q-value | 0.68186307 |
| FWER p-Value | 0.777 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Isg15 | 62 | 1.011 | 0.0209 | No |
| 2 | Tnfaip3 | 134 | 0.861 | 0.0378 | No |
| 3 | Cd74 | 447 | 0.673 | 0.0380 | No |
| 4 | Fgl2 | 714 | 0.588 | 0.0386 | No |
| 5 | Tnfaip6 | 837 | 0.562 | 0.0458 | No |
| 6 | Marchf1 | 889 | 0.547 | 0.0562 | No |
| 7 | Irf1 | 926 | 0.538 | 0.0672 | No |
| 8 | Cmklr1 | 1358 | 0.472 | 0.0567 | No |
| 9 | Myd88 | 2068 | 0.398 | 0.0304 | No |
| 10 | Ifit3 | 2125 | 0.395 | 0.0370 | No |
| 11 | Lats2 | 2361 | 0.374 | 0.0340 | No |
| 12 | Stat1 | 2395 | 0.370 | 0.0412 | No |
| 13 | Txnip | 3094 | 0.320 | 0.0136 | No |
| 14 | Cxcl10 | 3290 | 0.303 | 0.0110 | No |
| 15 | Nfkbia | 3465 | 0.291 | 0.0091 | No |
| 16 | Gch1 | 3504 | 0.288 | 0.0141 | No |
| 17 | Psme2 | 4052 | 0.260 | -0.0073 | No |
| 18 | Ripk2 | 4176 | 0.252 | -0.0075 | No |
| 19 | Psma2 | 4228 | 0.249 | -0.0042 | No |
| 20 | Nup93 | 4710 | 0.225 | -0.0231 | No |
| 21 | Ciita | 4775 | 0.223 | -0.0210 | No |
| 22 | H2-Aa | 4947 | 0.218 | -0.0244 | No |
| 23 | Cd38 | 5048 | 0.212 | -0.0244 | No |
| 24 | Hif1a | 5224 | 0.201 | -0.0285 | No |
| 25 | Trafd1 | 5238 | 0.200 | -0.0244 | No |
| 26 | Tap1 | 5342 | 0.195 | -0.0249 | No |
| 27 | Arl4a | 5382 | 0.193 | -0.0223 | No |
| 28 | Tapbp | 5443 | 0.189 | -0.0209 | No |
| 29 | Sod2 | 5735 | 0.174 | -0.0314 | No |
| 30 | Vcam1 | 6117 | 0.154 | -0.0469 | No |
| 31 | Cmpk2 | 6526 | 0.142 | -0.0641 | No |
| 32 | Gpr18 | 6634 | 0.138 | -0.0662 | No |
| 33 | Auts2 | 6791 | 0.129 | -0.0710 | No |
| 34 | Rnf213 | 6801 | 0.128 | -0.0684 | No |
| 35 | Cmtr1 | 6802 | 0.128 | -0.0654 | No |
| 36 | Ube2l6 | 6902 | 0.125 | -0.0674 | No |
| 37 | Batf2 | 7404 | 0.104 | -0.0901 | No |
| 38 | Zbp1 | 7437 | 0.103 | -0.0893 | No |
| 39 | Eif2ak2 | 7450 | 0.102 | -0.0875 | No |
| 40 | Cfb | 7486 | 0.100 | -0.0869 | No |
| 41 | Pnp | 7560 | 0.098 | -0.0882 | No |
| 42 | Ifi30 | 7595 | 0.096 | -0.0877 | No |
| 43 | Ptpn6 | 7640 | 0.093 | -0.0877 | No |
| 44 | Nampt | 7667 | 0.092 | -0.0868 | No |
| 45 | Slc25a28 | 7714 | 0.090 | -0.0870 | No |
| 46 | Pde4b | 7724 | 0.089 | -0.0853 | No |
| 47 | Isg20 | 8037 | 0.076 | -0.0992 | No |
| 48 | Lcp2 | 8151 | 0.072 | -0.1032 | No |
| 49 | Fcgr1 | 8362 | 0.063 | -0.1123 | No |
| 50 | Ifi44 | 8372 | 0.062 | -0.1112 | No |
| 51 | Ifit2 | 8765 | 0.047 | -0.1299 | No |
| 52 | Ptpn2 | 8878 | 0.043 | -0.1345 | No |
| 53 | Jak2 | 8983 | 0.039 | -0.1388 | No |
| 54 | Ogfr | 9192 | 0.030 | -0.1486 | No |
| 55 | Cd86 | 9398 | 0.023 | -0.1584 | No |
| 56 | Rapgef6 | 9526 | 0.018 | -0.1643 | No |
| 57 | Wars1 | 9531 | 0.018 | -0.1641 | No |
| 58 | Rnf31 | 9597 | 0.015 | -0.1670 | No |
| 59 | Irf9 | 9717 | 0.010 | -0.1728 | No |
| 60 | Fas | 9736 | 0.010 | -0.1734 | No |
| 61 | Cd69 | 9767 | 0.009 | -0.1747 | No |
| 62 | Parp14 | 9789 | 0.008 | -0.1756 | No |
| 63 | Samhd1 | 9897 | 0.004 | -0.1809 | No |
| 64 | Pfkp | 9901 | 0.004 | -0.1810 | No |
| 65 | Cd40 | 10003 | 0.000 | -0.1861 | No |
| 66 | St3gal5 | 10199 | -0.002 | -0.1958 | No |
| 67 | Usp18 | 10320 | -0.006 | -0.2017 | No |
| 68 | Eif4e3 | 10539 | -0.014 | -0.2124 | No |
| 69 | Znfx1 | 10755 | -0.022 | -0.2227 | No |
| 70 | Ptpn1 | 10871 | -0.026 | -0.2278 | No |
| 71 | Il7 | 10921 | -0.028 | -0.2296 | No |
| 72 | Isoc1 | 10957 | -0.030 | -0.2307 | No |
| 73 | Mthfd2 | 11019 | -0.033 | -0.2330 | No |
| 74 | Pla2g4a | 11600 | -0.056 | -0.2608 | No |
| 75 | Ripk1 | 11715 | -0.061 | -0.2651 | No |
| 76 | Plscr1 | 11760 | -0.062 | -0.2659 | No |
| 77 | Pnpt1 | 11930 | -0.069 | -0.2727 | No |
| 78 | Bst2 | 11944 | -0.070 | -0.2717 | No |
| 79 | Lap3 | 12071 | -0.075 | -0.2763 | No |
| 80 | Gbp3 | 12285 | -0.083 | -0.2851 | No |
| 81 | Pim1 | 12295 | -0.083 | -0.2835 | No |
| 82 | Nod1 | 12776 | -0.104 | -0.3052 | No |
| 83 | Oas3 | 12947 | -0.110 | -0.3112 | No |
| 84 | Bpgm | 12990 | -0.112 | -0.3106 | No |
| 85 | Psmb2 | 13025 | -0.114 | -0.3096 | No |
| 86 | Ncoa3 | 13156 | -0.117 | -0.3134 | No |
| 87 | Socs1 | 13349 | -0.124 | -0.3201 | No |
| 88 | P2ry14 | 13536 | -0.131 | -0.3264 | No |
| 89 | Rigi | 13645 | -0.135 | -0.3286 | Yes |
| 90 | Casp4 | 13675 | -0.137 | -0.3268 | Yes |
| 91 | Stat4 | 13738 | -0.140 | -0.3266 | Yes |
| 92 | Rbck1 | 13751 | -0.140 | -0.3239 | Yes |
| 93 | Sppl2a | 13753 | -0.140 | -0.3206 | Yes |
| 94 | Adar | 13819 | -0.144 | -0.3205 | Yes |
| 95 | Sri | 13862 | -0.146 | -0.3191 | Yes |
| 96 | Ifi35 | 13864 | -0.146 | -0.3157 | Yes |
| 97 | Trim21 | 13904 | -0.147 | -0.3142 | Yes |
| 98 | Irf8 | 13950 | -0.149 | -0.3129 | Yes |
| 99 | Irf2 | 14202 | -0.160 | -0.3218 | Yes |
| 100 | Nfkb1 | 14256 | -0.162 | -0.3206 | Yes |
| 101 | Bank1 | 14360 | -0.167 | -0.3218 | Yes |
| 102 | Lysmd2 | 14393 | -0.169 | -0.3194 | Yes |
| 103 | Rsad2 | 14398 | -0.169 | -0.3156 | Yes |
| 104 | Mvp | 14519 | -0.175 | -0.3175 | Yes |
| 105 | Il4ra | 14630 | -0.180 | -0.3187 | Yes |
| 106 | Pml | 14635 | -0.180 | -0.3147 | Yes |
| 107 | Stat3 | 14640 | -0.180 | -0.3106 | Yes |
| 108 | Dhx58 | 14642 | -0.180 | -0.3063 | Yes |
| 109 | Itgb7 | 14766 | -0.187 | -0.3081 | Yes |
| 110 | Psma3 | 14848 | -0.191 | -0.3076 | Yes |
| 111 | St8sia4 | 14976 | -0.198 | -0.3093 | Yes |
| 112 | Trim25 | 15019 | -0.200 | -0.3067 | Yes |
| 113 | Il18bp | 15139 | -0.206 | -0.3078 | Yes |
| 114 | Tor1b | 15240 | -0.210 | -0.3079 | Yes |
| 115 | Cfh | 15274 | -0.211 | -0.3045 | Yes |
| 116 | Ptgs2 | 15375 | -0.213 | -0.3045 | Yes |
| 117 | Socs3 | 15473 | -0.218 | -0.3042 | Yes |
| 118 | Arid5b | 15534 | -0.220 | -0.3020 | Yes |
| 119 | Irf4 | 15550 | -0.220 | -0.2975 | Yes |
| 120 | Casp7 | 15791 | -0.231 | -0.3041 | Yes |
| 121 | Psmb10 | 15901 | -0.237 | -0.3039 | Yes |
| 122 | Ifi27 | 15920 | -0.238 | -0.2992 | Yes |
| 123 | Oas2 | 15938 | -0.240 | -0.2944 | Yes |
| 124 | Stat2 | 16256 | -0.260 | -0.3042 | Yes |
| 125 | Mx2 | 16369 | -0.266 | -0.3035 | Yes |
| 126 | Sp110 | 16548 | -0.275 | -0.3059 | Yes |
| 127 | Tdrd7 | 16566 | -0.276 | -0.3002 | Yes |
| 128 | Casp8 | 16575 | -0.277 | -0.2940 | Yes |
| 129 | Il15ra | 16594 | -0.278 | -0.2884 | Yes |
| 130 | Ifitm2 | 16718 | -0.284 | -0.2878 | Yes |
| 131 | Helz2 | 17041 | -0.303 | -0.2968 | Yes |
| 132 | Psme1 | 17291 | -0.316 | -0.3019 | Yes |
| 133 | Btg1 | 17353 | -0.319 | -0.2973 | Yes |
| 134 | Samd9l | 17374 | -0.321 | -0.2907 | Yes |
| 135 | Irf7 | 17630 | -0.337 | -0.2955 | Yes |
| 136 | Selp | 17690 | -0.341 | -0.2904 | Yes |
| 137 | Irf5 | 17826 | -0.354 | -0.2888 | Yes |
| 138 | Il10ra | 17850 | -0.356 | -0.2815 | Yes |
| 139 | Cd274 | 17939 | -0.362 | -0.2773 | Yes |
| 140 | Psmb8 | 18044 | -0.370 | -0.2738 | Yes |
| 141 | Casp1 | 18054 | -0.371 | -0.2654 | Yes |
| 142 | Ifih1 | 18062 | -0.372 | -0.2569 | Yes |
| 143 | Peli1 | 18297 | -0.394 | -0.2594 | Yes |
| 144 | Epsti1 | 18357 | -0.399 | -0.2528 | Yes |
| 145 | Vamp8 | 18362 | -0.400 | -0.2436 | Yes |
| 146 | Nlrc5 | 18373 | -0.400 | -0.2346 | Yes |
| 147 | Trim14 | 18686 | -0.427 | -0.2401 | Yes |
| 148 | Ddx60 | 18737 | -0.434 | -0.2323 | Yes |
| 149 | Rtp4 | 18899 | -0.458 | -0.2296 | Yes |
| 150 | Tnfsf10 | 19090 | -0.483 | -0.2277 | Yes |
| 151 | Psmb9 | 19129 | -0.490 | -0.2179 | Yes |
| 152 | H2-DMa | 19240 | -0.515 | -0.2112 | Yes |
| 153 | Tnfaip2 | 19270 | -0.522 | -0.2003 | Yes |
| 154 | Il15 | 19360 | -0.537 | -0.1920 | Yes |
| 155 | Ifnar2 | 19421 | -0.551 | -0.1820 | Yes |
| 156 | Cdkn1a | 19559 | -0.586 | -0.1749 | Yes |
| 157 | Vamp5 | 19598 | -0.596 | -0.1627 | Yes |
| 158 | Icam1 | 19617 | -0.601 | -0.1493 | Yes |
| 159 | Oasl1 | 19620 | -0.603 | -0.1351 | Yes |
| 160 | Nmi | 19627 | -0.606 | -0.1210 | Yes |
| 161 | Xaf1 | 19637 | -0.608 | -0.1070 | Yes |
| 162 | Trim26 | 19665 | -0.621 | -0.0936 | Yes |
| 163 | B2m | 19702 | -0.634 | -0.0804 | Yes |
| 164 | Lgals3bp | 19757 | -0.653 | -0.0676 | Yes |
| 165 | Parp12 | 19763 | -0.655 | -0.0523 | Yes |
| 166 | Casp3 | 19766 | -0.656 | -0.0368 | Yes |
| 167 | Ly6e | 19792 | -0.667 | -0.0222 | Yes |
| 168 | Herc6 | 19796 | -0.671 | -0.0064 | Yes |
| 169 | Ifitm3 | 19896 | -0.756 | 0.0065 | Yes |