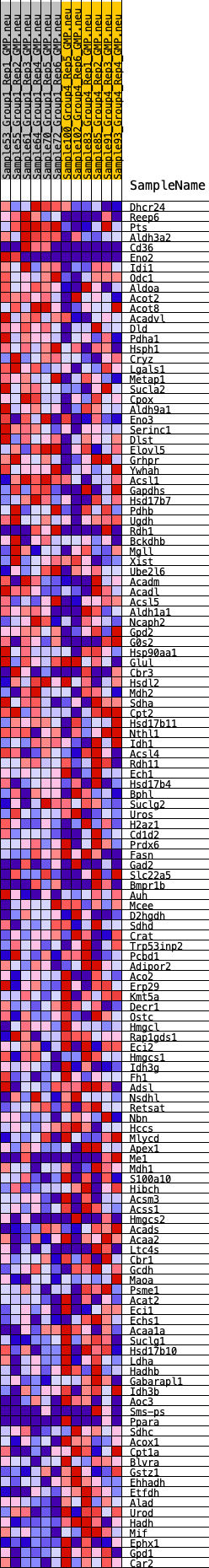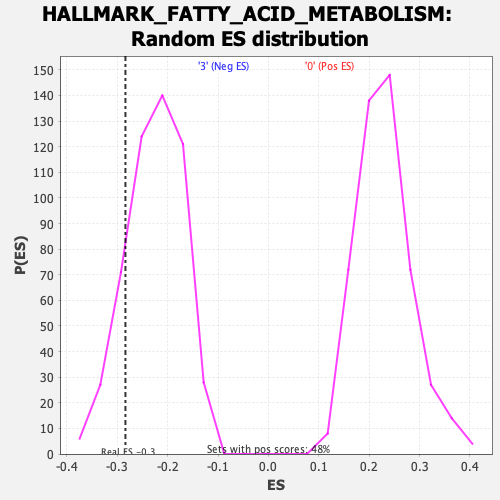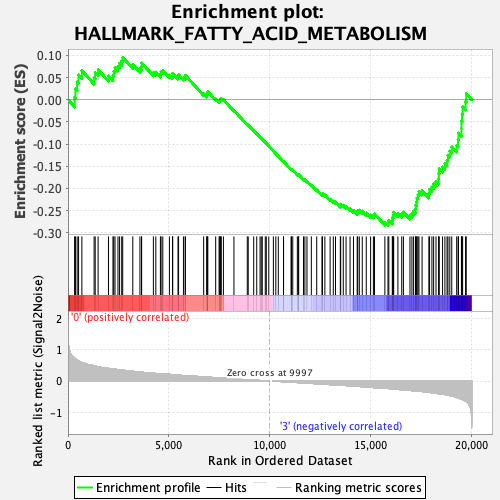
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | GMP.GMP.neu_Pheno.cls #Group1_versus_Group4.GMP.neu_Pheno.cls #Group1_versus_Group4_repos |
| Phenotype | GMP.neu_Pheno.cls#Group1_versus_Group4_repos |
| Upregulated in class | 3 |
| GeneSet | HALLMARK_FATTY_ACID_METABOLISM |
| Enrichment Score (ES) | -0.28377092 |
| Normalized Enrichment Score (NES) | -1.2473035 |
| Nominal p-value | 0.15860735 |
| FDR q-value | 0.73995143 |
| FWER p-Value | 0.868 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Dhcr24 | 327 | 0.729 | 0.0054 | No |
| 2 | Reep6 | 368 | 0.711 | 0.0247 | No |
| 3 | Pts | 454 | 0.669 | 0.0404 | No |
| 4 | Aldh3a2 | 524 | 0.641 | 0.0561 | No |
| 5 | Cd36 | 688 | 0.590 | 0.0656 | No |
| 6 | Eno2 | 1295 | 0.485 | 0.0497 | No |
| 7 | Idi1 | 1349 | 0.473 | 0.0612 | No |
| 8 | Odc1 | 1493 | 0.453 | 0.0675 | No |
| 9 | Aldoa | 2007 | 0.404 | 0.0539 | No |
| 10 | Acot2 | 2230 | 0.386 | 0.0542 | No |
| 11 | Acot8 | 2269 | 0.383 | 0.0638 | No |
| 12 | Acadvl | 2331 | 0.377 | 0.0720 | No |
| 13 | Dld | 2478 | 0.363 | 0.0755 | No |
| 14 | Pdha1 | 2558 | 0.357 | 0.0823 | No |
| 15 | Hsph1 | 2663 | 0.351 | 0.0875 | No |
| 16 | Cryz | 2713 | 0.345 | 0.0954 | No |
| 17 | Lgals1 | 3214 | 0.309 | 0.0795 | No |
| 18 | Metap1 | 3567 | 0.284 | 0.0703 | No |
| 19 | Sucla2 | 3646 | 0.281 | 0.0748 | No |
| 20 | Cpox | 3652 | 0.281 | 0.0830 | No |
| 21 | Aldh9a1 | 4234 | 0.248 | 0.0612 | No |
| 22 | Eno3 | 4355 | 0.240 | 0.0624 | No |
| 23 | Serinc1 | 4582 | 0.232 | 0.0579 | No |
| 24 | Dlst | 4616 | 0.230 | 0.0632 | No |
| 25 | Elovl5 | 4701 | 0.225 | 0.0657 | No |
| 26 | Grhpr | 5029 | 0.213 | 0.0556 | No |
| 27 | Ywhah | 5183 | 0.204 | 0.0540 | No |
| 28 | Acsl1 | 5193 | 0.203 | 0.0597 | No |
| 29 | Gapdhs | 5456 | 0.188 | 0.0521 | No |
| 30 | Hsd17b7 | 5484 | 0.187 | 0.0564 | No |
| 31 | Pdhb | 5725 | 0.174 | 0.0495 | No |
| 32 | Ugdh | 5813 | 0.170 | 0.0502 | No |
| 33 | Rdh1 | 5818 | 0.169 | 0.0551 | No |
| 34 | Bckdhb | 6728 | 0.133 | 0.0133 | No |
| 35 | Mgll | 6883 | 0.126 | 0.0094 | No |
| 36 | Xist | 6898 | 0.126 | 0.0124 | No |
| 37 | Ube2l6 | 6902 | 0.125 | 0.0160 | No |
| 38 | Acadm | 6934 | 0.124 | 0.0182 | No |
| 39 | Acadl | 7324 | 0.107 | 0.0018 | No |
| 40 | Acsl5 | 7493 | 0.100 | -0.0036 | No |
| 41 | Aldh1a1 | 7529 | 0.099 | -0.0024 | No |
| 42 | Ncaph2 | 7542 | 0.099 | -0.0001 | No |
| 43 | Gpd2 | 7570 | 0.097 | 0.0015 | No |
| 44 | G0s2 | 7600 | 0.096 | 0.0029 | No |
| 45 | Hsp90aa1 | 7707 | 0.090 | 0.0003 | No |
| 46 | Glul | 8230 | 0.068 | -0.0239 | No |
| 47 | Cbr3 | 8890 | 0.043 | -0.0558 | No |
| 48 | Hsdl2 | 8938 | 0.040 | -0.0569 | No |
| 49 | Mdh2 | 9210 | 0.029 | -0.0697 | No |
| 50 | Sdha | 9360 | 0.024 | -0.0765 | No |
| 51 | Cpt2 | 9525 | 0.018 | -0.0842 | No |
| 52 | Hsd17b11 | 9583 | 0.016 | -0.0866 | No |
| 53 | Nthl1 | 9635 | 0.014 | -0.0887 | No |
| 54 | Idh1 | 9783 | 0.008 | -0.0959 | No |
| 55 | Acsl4 | 9834 | 0.006 | -0.0982 | No |
| 56 | Rdh11 | 9957 | 0.002 | -0.1043 | No |
| 57 | Ech1 | 10185 | -0.001 | -0.1156 | No |
| 58 | Hsd17b4 | 10306 | -0.006 | -0.1215 | No |
| 59 | Bphl | 10429 | -0.010 | -0.1273 | No |
| 60 | Suclg2 | 10688 | -0.020 | -0.1397 | No |
| 61 | Uros | 10689 | -0.020 | -0.1391 | No |
| 62 | H2az1 | 11062 | -0.035 | -0.1567 | No |
| 63 | Cd1d2 | 11085 | -0.036 | -0.1568 | No |
| 64 | Prdx6 | 11146 | -0.038 | -0.1586 | No |
| 65 | Fasn | 11373 | -0.047 | -0.1686 | No |
| 66 | Gad2 | 11426 | -0.049 | -0.1697 | No |
| 67 | Slc22a5 | 11439 | -0.050 | -0.1689 | No |
| 68 | Bmpr1b | 11679 | -0.059 | -0.1791 | No |
| 69 | Auh | 11748 | -0.062 | -0.1807 | No |
| 70 | Mcee | 11851 | -0.067 | -0.1838 | No |
| 71 | D2hgdh | 12067 | -0.075 | -0.1924 | No |
| 72 | Sdhd | 12335 | -0.085 | -0.2032 | No |
| 73 | Crat | 12595 | -0.096 | -0.2134 | No |
| 74 | Trp53inp2 | 12620 | -0.098 | -0.2116 | No |
| 75 | Pcbd1 | 12739 | -0.103 | -0.2145 | No |
| 76 | Adipor2 | 12999 | -0.113 | -0.2241 | No |
| 77 | Aco2 | 13157 | -0.117 | -0.2285 | No |
| 78 | Erp29 | 13266 | -0.121 | -0.2303 | No |
| 79 | Kmt5a | 13504 | -0.130 | -0.2384 | No |
| 80 | Decr1 | 13524 | -0.131 | -0.2354 | No |
| 81 | Ostc | 13652 | -0.136 | -0.2377 | No |
| 82 | Hmgcl | 13789 | -0.143 | -0.2403 | No |
| 83 | Rap1gds1 | 13988 | -0.151 | -0.2458 | No |
| 84 | Eci2 | 14157 | -0.158 | -0.2495 | No |
| 85 | Hmgcs1 | 14342 | -0.166 | -0.2537 | No |
| 86 | Idh3g | 14369 | -0.167 | -0.2500 | No |
| 87 | Fh1 | 14447 | -0.172 | -0.2488 | No |
| 88 | Adsl | 14593 | -0.178 | -0.2507 | No |
| 89 | Nsdhl | 14792 | -0.188 | -0.2550 | No |
| 90 | Retsat | 15007 | -0.199 | -0.2598 | No |
| 91 | Nbn | 15145 | -0.206 | -0.2606 | No |
| 92 | Hccs | 15201 | -0.208 | -0.2571 | No |
| 93 | Mlycd | 15714 | -0.228 | -0.2760 | No |
| 94 | Apex1 | 15869 | -0.236 | -0.2767 | Yes |
| 95 | Me1 | 15910 | -0.238 | -0.2716 | Yes |
| 96 | Mdh1 | 16083 | -0.249 | -0.2728 | Yes |
| 97 | S100a10 | 16092 | -0.250 | -0.2657 | Yes |
| 98 | Hibch | 16133 | -0.252 | -0.2602 | Yes |
| 99 | Acsm3 | 16151 | -0.254 | -0.2534 | Yes |
| 100 | Acss1 | 16361 | -0.266 | -0.2560 | Yes |
| 101 | Hmgcs2 | 16551 | -0.275 | -0.2573 | Yes |
| 102 | Acads | 16634 | -0.279 | -0.2530 | Yes |
| 103 | Acaa2 | 16959 | -0.298 | -0.2604 | Yes |
| 104 | Ltc4s | 17056 | -0.303 | -0.2561 | Yes |
| 105 | Cbr1 | 17139 | -0.308 | -0.2510 | Yes |
| 106 | Gcdh | 17233 | -0.314 | -0.2463 | Yes |
| 107 | Maoa | 17247 | -0.315 | -0.2376 | Yes |
| 108 | Psme1 | 17291 | -0.316 | -0.2303 | Yes |
| 109 | Acat2 | 17307 | -0.317 | -0.2215 | Yes |
| 110 | Eci1 | 17358 | -0.320 | -0.2145 | Yes |
| 111 | Echs1 | 17405 | -0.323 | -0.2071 | Yes |
| 112 | Acaa1a | 17560 | -0.332 | -0.2049 | Yes |
| 113 | Suclg1 | 17897 | -0.359 | -0.2110 | Yes |
| 114 | Hsd17b10 | 17931 | -0.362 | -0.2019 | Yes |
| 115 | Ldha | 18048 | -0.371 | -0.1966 | Yes |
| 116 | Hadhb | 18132 | -0.378 | -0.1895 | Yes |
| 117 | Gabarapl1 | 18250 | -0.390 | -0.1837 | Yes |
| 118 | Idh3b | 18381 | -0.401 | -0.1782 | Yes |
| 119 | Aoc3 | 18388 | -0.402 | -0.1665 | Yes |
| 120 | Sms-ps | 18407 | -0.403 | -0.1553 | Yes |
| 121 | Ppara | 18576 | -0.418 | -0.1513 | Yes |
| 122 | Sdhc | 18692 | -0.428 | -0.1442 | Yes |
| 123 | Acox1 | 18798 | -0.442 | -0.1363 | Yes |
| 124 | Cpt1a | 18843 | -0.451 | -0.1250 | Yes |
| 125 | Blvra | 18935 | -0.463 | -0.1157 | Yes |
| 126 | Gstz1 | 19031 | -0.475 | -0.1063 | Yes |
| 127 | Ehhadh | 19276 | -0.523 | -0.1029 | Yes |
| 128 | Etfdh | 19347 | -0.535 | -0.0904 | Yes |
| 129 | Alad | 19365 | -0.538 | -0.0752 | Yes |
| 130 | Urod | 19512 | -0.574 | -0.0653 | Yes |
| 131 | Hadh | 19513 | -0.574 | -0.0481 | Yes |
| 132 | Mif | 19544 | -0.582 | -0.0322 | Yes |
| 133 | Ephx1 | 19571 | -0.589 | -0.0159 | Yes |
| 134 | Gpd1 | 19716 | -0.639 | -0.0040 | Yes |
| 135 | Car2 | 19749 | -0.652 | 0.0139 | Yes |