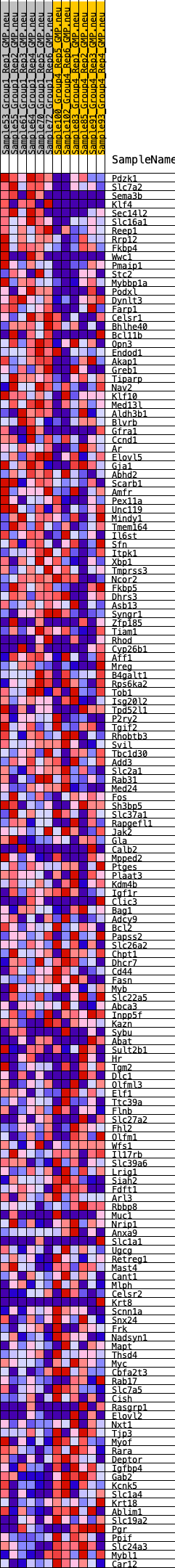
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | GMP.GMP.neu_Pheno.cls #Group1_versus_Group4.GMP.neu_Pheno.cls #Group1_versus_Group4_repos |
| Phenotype | GMP.neu_Pheno.cls#Group1_versus_Group4_repos |
| Upregulated in class | 0 |
| GeneSet | HALLMARK_ESTROGEN_RESPONSE_EARLY |
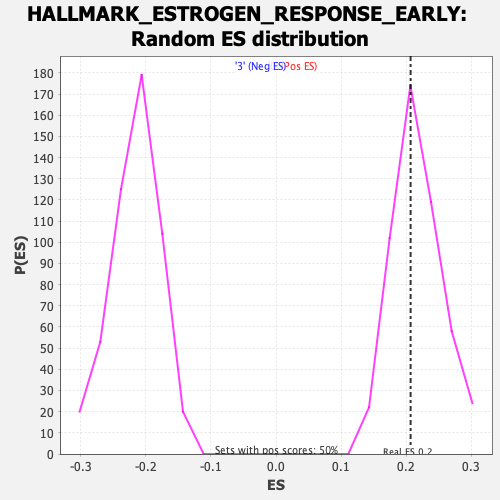
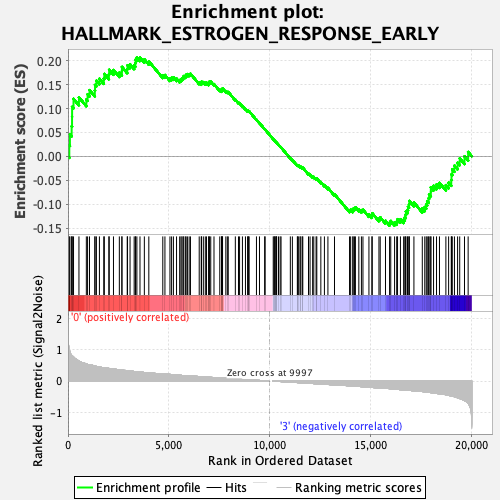
| Enrichment Score (ES) | 0.20686877 |
| Normalized Enrichment Score (NES) | 0.9560633 |
| Nominal p-value | 0.5591182 |
| FDR q-value | 1.0 |
| FWER p-Value | 0.998 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Pdzk1 | 65 | 1.004 | 0.0229 | Yes |
| 2 | Slc7a2 | 95 | 0.934 | 0.0457 | Yes |
| 3 | Sema3b | 181 | 0.818 | 0.0628 | Yes |
| 4 | Klf4 | 201 | 0.806 | 0.0828 | Yes |
| 5 | Sec14l2 | 204 | 0.805 | 0.1037 | Yes |
| 6 | Slc16a1 | 274 | 0.766 | 0.1202 | Yes |
| 7 | Reep1 | 543 | 0.636 | 0.1232 | Yes |
| 8 | Rrp12 | 909 | 0.543 | 0.1190 | Yes |
| 9 | Fkbp4 | 971 | 0.529 | 0.1297 | Yes |
| 10 | Wwc1 | 1066 | 0.514 | 0.1384 | Yes |
| 11 | Pmaip1 | 1330 | 0.478 | 0.1376 | Yes |
| 12 | Stc2 | 1341 | 0.475 | 0.1495 | Yes |
| 13 | Mybbp1a | 1409 | 0.465 | 0.1582 | Yes |
| 14 | Podxl | 1558 | 0.445 | 0.1623 | Yes |
| 15 | Dynlt3 | 1774 | 0.425 | 0.1626 | Yes |
| 16 | Farp1 | 1801 | 0.423 | 0.1723 | Yes |
| 17 | Celsr1 | 2027 | 0.402 | 0.1715 | Yes |
| 18 | Bhlhe40 | 2042 | 0.400 | 0.1812 | Yes |
| 19 | Bcl11b | 2255 | 0.384 | 0.1805 | Yes |
| 20 | Opn3 | 2546 | 0.359 | 0.1752 | Yes |
| 21 | Endod1 | 2672 | 0.350 | 0.1780 | Yes |
| 22 | Akap1 | 2675 | 0.349 | 0.1870 | Yes |
| 23 | Greb1 | 2937 | 0.331 | 0.1825 | Yes |
| 24 | Tiparp | 2952 | 0.330 | 0.1904 | Yes |
| 25 | Nav2 | 3077 | 0.322 | 0.1926 | Yes |
| 26 | Klf10 | 3281 | 0.304 | 0.1903 | Yes |
| 27 | Med13l | 3341 | 0.300 | 0.1951 | Yes |
| 28 | Aldh3b1 | 3352 | 0.299 | 0.2024 | Yes |
| 29 | Blvrb | 3416 | 0.294 | 0.2069 | Yes |
| 30 | Gfra1 | 3570 | 0.284 | 0.2066 | No |
| 31 | Ccnd1 | 3786 | 0.272 | 0.2028 | No |
| 32 | Ar | 4008 | 0.262 | 0.1985 | No |
| 33 | Elovl5 | 4701 | 0.225 | 0.1696 | No |
| 34 | Gja1 | 4807 | 0.222 | 0.1701 | No |
| 35 | Abhd2 | 5056 | 0.211 | 0.1631 | No |
| 36 | Scarb1 | 5138 | 0.206 | 0.1644 | No |
| 37 | Amfr | 5227 | 0.201 | 0.1652 | No |
| 38 | Pex11a | 5383 | 0.193 | 0.1624 | No |
| 39 | Unc119 | 5538 | 0.184 | 0.1594 | No |
| 40 | Mindy1 | 5601 | 0.181 | 0.1610 | No |
| 41 | Tmem164 | 5651 | 0.178 | 0.1632 | No |
| 42 | Il6st | 5708 | 0.175 | 0.1649 | No |
| 43 | Sfn | 5740 | 0.173 | 0.1679 | No |
| 44 | Itpk1 | 5820 | 0.169 | 0.1683 | No |
| 45 | Xbp1 | 5857 | 0.168 | 0.1709 | No |
| 46 | Tmprss3 | 5922 | 0.165 | 0.1719 | No |
| 47 | Ncor2 | 6008 | 0.160 | 0.1718 | No |
| 48 | Fkbp5 | 6073 | 0.156 | 0.1727 | No |
| 49 | Dhrs3 | 6507 | 0.143 | 0.1546 | No |
| 50 | Asb13 | 6607 | 0.139 | 0.1532 | No |
| 51 | Syngr1 | 6621 | 0.138 | 0.1562 | No |
| 52 | Zfp185 | 6711 | 0.133 | 0.1552 | No |
| 53 | Tiam1 | 6808 | 0.128 | 0.1537 | No |
| 54 | Rhod | 6867 | 0.127 | 0.1540 | No |
| 55 | Cyp26b1 | 6974 | 0.123 | 0.1519 | No |
| 56 | Aff1 | 6992 | 0.121 | 0.1542 | No |
| 57 | Mreg | 7012 | 0.120 | 0.1564 | No |
| 58 | B4galt1 | 7079 | 0.117 | 0.1561 | No |
| 59 | Rps6ka2 | 7240 | 0.110 | 0.1509 | No |
| 60 | Tob1 | 7527 | 0.099 | 0.1391 | No |
| 61 | Isg20l2 | 7612 | 0.095 | 0.1374 | No |
| 62 | Tpd52l1 | 7618 | 0.095 | 0.1396 | No |
| 63 | P2ry2 | 7657 | 0.092 | 0.1401 | No |
| 64 | Tgif2 | 7664 | 0.092 | 0.1422 | No |
| 65 | Rhobtb3 | 7827 | 0.085 | 0.1362 | No |
| 66 | Svil | 7902 | 0.081 | 0.1346 | No |
| 67 | Tbc1d30 | 7947 | 0.080 | 0.1345 | No |
| 68 | Add3 | 8298 | 0.065 | 0.1186 | No |
| 69 | Slc2a1 | 8455 | 0.059 | 0.1122 | No |
| 70 | Rab31 | 8509 | 0.056 | 0.1110 | No |
| 71 | Med24 | 8650 | 0.050 | 0.1053 | No |
| 72 | Fos | 8800 | 0.045 | 0.0990 | No |
| 73 | Sh3bp5 | 8909 | 0.042 | 0.0946 | No |
| 74 | Slc37a1 | 8934 | 0.040 | 0.0945 | No |
| 75 | Rapgefl1 | 8941 | 0.040 | 0.0952 | No |
| 76 | Jak2 | 8983 | 0.039 | 0.0942 | No |
| 77 | Gla | 9346 | 0.024 | 0.0766 | No |
| 78 | Calb2 | 9496 | 0.019 | 0.0696 | No |
| 79 | Mpped2 | 9752 | 0.009 | 0.0570 | No |
| 80 | Ptges | 9776 | 0.008 | 0.0560 | No |
| 81 | Plaat3 | 10178 | -0.001 | 0.0359 | No |
| 82 | Kdm4b | 10217 | -0.003 | 0.0340 | No |
| 83 | Igf1r | 10249 | -0.004 | 0.0326 | No |
| 84 | Clic3 | 10271 | -0.005 | 0.0316 | No |
| 85 | Bag1 | 10312 | -0.006 | 0.0298 | No |
| 86 | Adcy9 | 10335 | -0.007 | 0.0288 | No |
| 87 | Bcl2 | 10348 | -0.008 | 0.0284 | No |
| 88 | Papss2 | 10439 | -0.011 | 0.0242 | No |
| 89 | Slc26a2 | 10519 | -0.013 | 0.0205 | No |
| 90 | Chpt1 | 10565 | -0.015 | 0.0187 | No |
| 91 | Dhcr7 | 11026 | -0.033 | -0.0036 | No |
| 92 | Cd44 | 11127 | -0.038 | -0.0077 | No |
| 93 | Fasn | 11373 | -0.047 | -0.0188 | No |
| 94 | Myb | 11389 | -0.048 | -0.0183 | No |
| 95 | Slc22a5 | 11439 | -0.050 | -0.0195 | No |
| 96 | Abca3 | 11508 | -0.053 | -0.0215 | No |
| 97 | Inpp5f | 11544 | -0.054 | -0.0219 | No |
| 98 | Kazn | 11618 | -0.057 | -0.0241 | No |
| 99 | Sybu | 11644 | -0.058 | -0.0238 | No |
| 100 | Abat | 11927 | -0.069 | -0.0362 | No |
| 101 | Sult2b1 | 12004 | -0.072 | -0.0381 | No |
| 102 | Hr | 12132 | -0.078 | -0.0425 | No |
| 103 | Tgm2 | 12189 | -0.080 | -0.0433 | No |
| 104 | Dlc1 | 12303 | -0.083 | -0.0468 | No |
| 105 | Olfml3 | 12339 | -0.085 | -0.0463 | No |
| 106 | Elf1 | 12539 | -0.094 | -0.0539 | No |
| 107 | Ttc39a | 12716 | -0.102 | -0.0601 | No |
| 108 | Flnb | 12890 | -0.108 | -0.0660 | No |
| 109 | Slc27a2 | 13216 | -0.119 | -0.0792 | No |
| 110 | Fhl2 | 13962 | -0.150 | -0.1128 | No |
| 111 | Olfm1 | 14017 | -0.152 | -0.1116 | No |
| 112 | Wfs1 | 14123 | -0.157 | -0.1128 | No |
| 113 | Il17rb | 14141 | -0.158 | -0.1095 | No |
| 114 | Slc39a6 | 14206 | -0.160 | -0.1086 | No |
| 115 | Lrig1 | 14253 | -0.162 | -0.1067 | No |
| 116 | Siah2 | 14423 | -0.171 | -0.1107 | No |
| 117 | Fdft1 | 14551 | -0.176 | -0.1126 | No |
| 118 | Arl3 | 14627 | -0.180 | -0.1116 | No |
| 119 | Rbbp8 | 14924 | -0.195 | -0.1215 | No |
| 120 | Muc1 | 15070 | -0.202 | -0.1235 | No |
| 121 | Nrip1 | 15087 | -0.203 | -0.1190 | No |
| 122 | Anxa9 | 15421 | -0.216 | -0.1302 | No |
| 123 | Slc1a1 | 15483 | -0.219 | -0.1275 | No |
| 124 | Ugcg | 15745 | -0.229 | -0.1347 | No |
| 125 | Retreg1 | 15947 | -0.240 | -0.1386 | No |
| 126 | Mast4 | 16006 | -0.244 | -0.1351 | No |
| 127 | Cant1 | 16196 | -0.257 | -0.1380 | No |
| 128 | Mlph | 16301 | -0.263 | -0.1363 | No |
| 129 | Celsr2 | 16343 | -0.265 | -0.1315 | No |
| 130 | Krt8 | 16486 | -0.272 | -0.1316 | No |
| 131 | Scnn1a | 16645 | -0.280 | -0.1322 | No |
| 132 | Snx24 | 16703 | -0.284 | -0.1277 | No |
| 133 | Frk | 16743 | -0.285 | -0.1223 | No |
| 134 | Nadsyn1 | 16757 | -0.286 | -0.1154 | No |
| 135 | Mapt | 16837 | -0.291 | -0.1119 | No |
| 136 | Thsd4 | 16863 | -0.292 | -0.1055 | No |
| 137 | Myc | 16912 | -0.295 | -0.1002 | No |
| 138 | Cbfa2t3 | 16928 | -0.296 | -0.0932 | No |
| 139 | Rab17 | 17155 | -0.309 | -0.0966 | No |
| 140 | Slc7a5 | 17570 | -0.333 | -0.1087 | No |
| 141 | Cish | 17693 | -0.342 | -0.1060 | No |
| 142 | Rasgrp1 | 17766 | -0.349 | -0.1005 | No |
| 143 | Elovl2 | 17815 | -0.353 | -0.0937 | No |
| 144 | Nxt1 | 17883 | -0.357 | -0.0878 | No |
| 145 | Tjp3 | 17909 | -0.360 | -0.0797 | No |
| 146 | Myof | 17993 | -0.367 | -0.0743 | No |
| 147 | Rara | 17994 | -0.367 | -0.0647 | No |
| 148 | Deptor | 18129 | -0.378 | -0.0616 | No |
| 149 | Igfbp4 | 18276 | -0.392 | -0.0588 | No |
| 150 | Gab2 | 18422 | -0.404 | -0.0556 | No |
| 151 | Kcnk5 | 18740 | -0.434 | -0.0602 | No |
| 152 | Slc1a4 | 18876 | -0.455 | -0.0551 | No |
| 153 | Krt18 | 19007 | -0.471 | -0.0494 | No |
| 154 | Ablim1 | 19019 | -0.474 | -0.0376 | No |
| 155 | Slc19a2 | 19057 | -0.478 | -0.0270 | No |
| 156 | Pgr | 19164 | -0.497 | -0.0194 | No |
| 157 | Ppif | 19321 | -0.529 | -0.0135 | No |
| 158 | Slc24a3 | 19426 | -0.554 | -0.0043 | No |
| 159 | Mybl1 | 19663 | -0.621 | 0.0000 | No |
| 160 | Car12 | 19844 | -0.699 | 0.0092 | No |