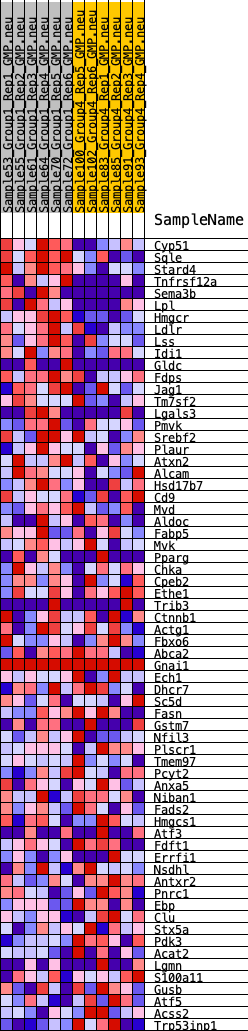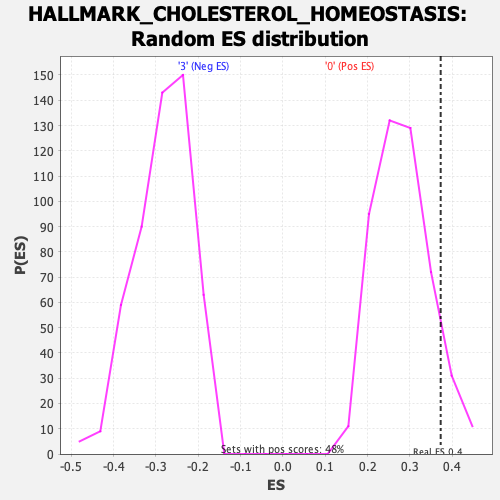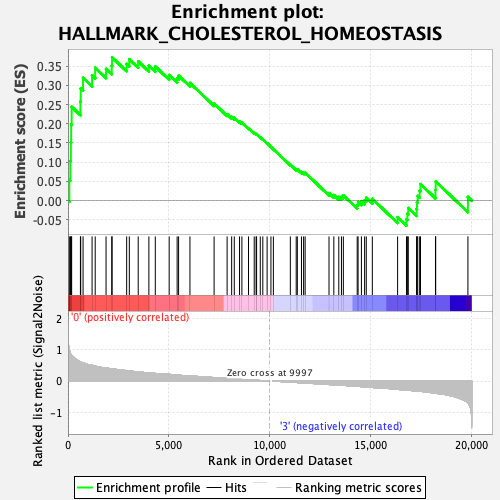
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | GMP.GMP.neu_Pheno.cls #Group1_versus_Group4.GMP.neu_Pheno.cls #Group1_versus_Group4_repos |
| Phenotype | GMP.neu_Pheno.cls#Group1_versus_Group4_repos |
| Upregulated in class | 0 |
| GeneSet | HALLMARK_CHOLESTEROL_HOMEOSTASIS |
| Enrichment Score (ES) | 0.37272662 |
| Normalized Enrichment Score (NES) | 1.3210967 |
| Nominal p-value | 0.0977131 |
| FDR q-value | 0.6899846 |
| FWER p-Value | 0.73 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Cyp51 | 66 | 1.002 | 0.0543 | Yes |
| 2 | Sqle | 117 | 0.898 | 0.1034 | Yes |
| 3 | Stard4 | 141 | 0.853 | 0.1513 | Yes |
| 4 | Tnfrsf12a | 158 | 0.829 | 0.1982 | Yes |
| 5 | Sema3b | 181 | 0.818 | 0.2441 | Yes |
| 6 | Lpl | 616 | 0.613 | 0.2576 | Yes |
| 7 | Hmgcr | 630 | 0.608 | 0.2919 | Yes |
| 8 | Ldlr | 747 | 0.582 | 0.3196 | Yes |
| 9 | Lss | 1194 | 0.505 | 0.3263 | Yes |
| 10 | Idi1 | 1349 | 0.473 | 0.3458 | Yes |
| 11 | Gldc | 1887 | 0.416 | 0.3427 | Yes |
| 12 | Fdps | 2170 | 0.391 | 0.3511 | Yes |
| 13 | Jag1 | 2186 | 0.389 | 0.3727 | Yes |
| 14 | Tm7sf2 | 2909 | 0.334 | 0.3557 | No |
| 15 | Lgals3 | 3041 | 0.324 | 0.3678 | No |
| 16 | Pmvk | 3482 | 0.290 | 0.3624 | No |
| 17 | Srebf2 | 4011 | 0.262 | 0.3511 | No |
| 18 | Plaur | 4329 | 0.242 | 0.3491 | No |
| 19 | Atxn2 | 5017 | 0.213 | 0.3269 | No |
| 20 | Alcam | 5412 | 0.191 | 0.3182 | No |
| 21 | Hsd17b7 | 5484 | 0.187 | 0.3254 | No |
| 22 | Cd9 | 6048 | 0.157 | 0.3062 | No |
| 23 | Mvd | 7245 | 0.110 | 0.2526 | No |
| 24 | Aldoc | 7891 | 0.082 | 0.2250 | No |
| 25 | Fabp5 | 8116 | 0.073 | 0.2180 | No |
| 26 | Mvk | 8246 | 0.067 | 0.2154 | No |
| 27 | Pparg | 8507 | 0.056 | 0.2056 | No |
| 28 | Chka | 8624 | 0.051 | 0.2027 | No |
| 29 | Cpeb2 | 8952 | 0.040 | 0.1886 | No |
| 30 | Ethe1 | 9235 | 0.028 | 0.1761 | No |
| 31 | Trib3 | 9328 | 0.024 | 0.1729 | No |
| 32 | Ctnnb1 | 9353 | 0.024 | 0.1731 | No |
| 33 | Actg1 | 9535 | 0.018 | 0.1651 | No |
| 34 | Fbxo6 | 9661 | 0.012 | 0.1595 | No |
| 35 | Abca2 | 9874 | 0.005 | 0.1492 | No |
| 36 | Gnai1 | 10064 | 0.000 | 0.1397 | No |
| 37 | Ech1 | 10185 | -0.001 | 0.1338 | No |
| 38 | Dhcr7 | 11026 | -0.033 | 0.0936 | No |
| 39 | Sc5d | 11317 | -0.045 | 0.0816 | No |
| 40 | Fasn | 11373 | -0.047 | 0.0816 | No |
| 41 | Gstm7 | 11578 | -0.056 | 0.0746 | No |
| 42 | Nfil3 | 11684 | -0.059 | 0.0727 | No |
| 43 | Plscr1 | 11760 | -0.062 | 0.0725 | No |
| 44 | Tmem97 | 12944 | -0.110 | 0.0196 | No |
| 45 | Pcyt2 | 13183 | -0.118 | 0.0145 | No |
| 46 | Anxa5 | 13429 | -0.127 | 0.0095 | No |
| 47 | Niban1 | 13561 | -0.132 | 0.0105 | No |
| 48 | Fads2 | 13655 | -0.136 | 0.0137 | No |
| 49 | Hmgcs1 | 14342 | -0.166 | -0.0111 | No |
| 50 | Atf3 | 14386 | -0.168 | -0.0036 | No |
| 51 | Fdft1 | 14551 | -0.176 | -0.0017 | No |
| 52 | Errfi1 | 14710 | -0.184 | 0.0010 | No |
| 53 | Nsdhl | 14792 | -0.188 | 0.0077 | No |
| 54 | Antxr2 | 15092 | -0.203 | 0.0044 | No |
| 55 | Pnrc1 | 16346 | -0.265 | -0.0431 | No |
| 56 | Ebp | 16786 | -0.288 | -0.0485 | No |
| 57 | Clu | 16831 | -0.290 | -0.0340 | No |
| 58 | Stx5a | 16876 | -0.293 | -0.0194 | No |
| 59 | Pdk3 | 17287 | -0.316 | -0.0218 | No |
| 60 | Acat2 | 17307 | -0.317 | -0.0045 | No |
| 61 | Lgmn | 17344 | -0.319 | 0.0121 | No |
| 62 | S100a11 | 17440 | -0.326 | 0.0260 | No |
| 63 | Gusb | 17481 | -0.328 | 0.0429 | No |
| 64 | Atf5 | 18225 | -0.387 | 0.0279 | No |
| 65 | Acss2 | 18235 | -0.388 | 0.0498 | No |
| 66 | Trp53inp1 | 19833 | -0.694 | 0.0097 | No |