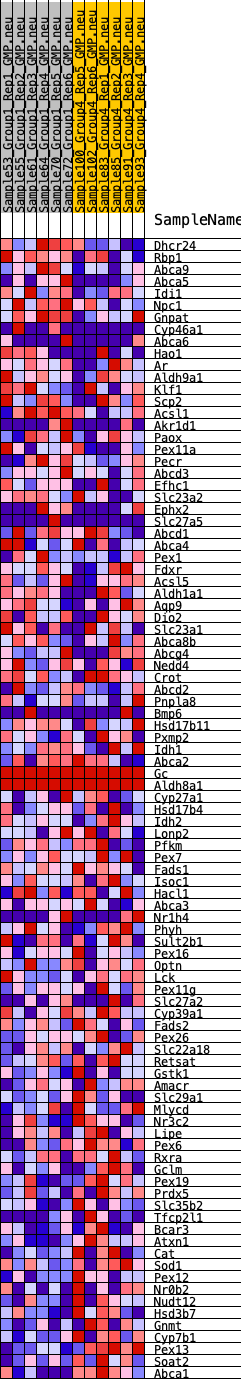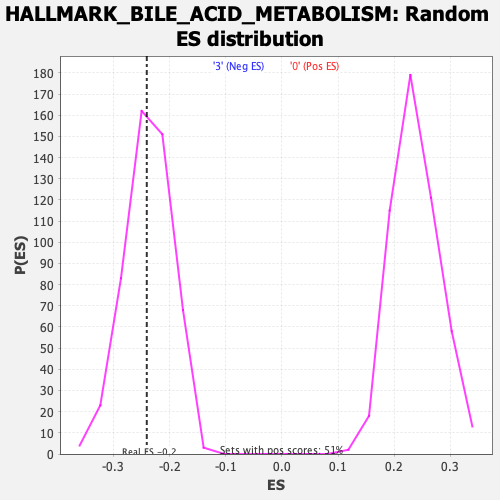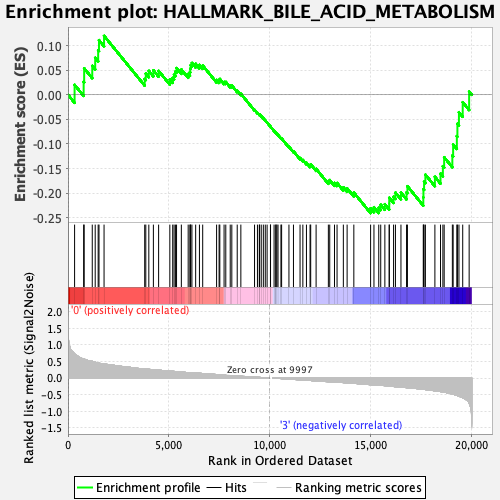
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | GMP.GMP.neu_Pheno.cls #Group1_versus_Group4.GMP.neu_Pheno.cls #Group1_versus_Group4_repos |
| Phenotype | GMP.neu_Pheno.cls#Group1_versus_Group4_repos |
| Upregulated in class | 3 |
| GeneSet | HALLMARK_BILE_ACID_METABOLISM |
| Enrichment Score (ES) | -0.24051559 |
| Normalized Enrichment Score (NES) | -1.0105017 |
| Nominal p-value | 0.46153846 |
| FDR q-value | 0.73448867 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Dhcr24 | 327 | 0.729 | 0.0201 | No |
| 2 | Rbp1 | 776 | 0.575 | 0.0264 | No |
| 3 | Abca9 | 802 | 0.570 | 0.0537 | No |
| 4 | Abca5 | 1203 | 0.503 | 0.0588 | No |
| 5 | Idi1 | 1349 | 0.473 | 0.0753 | No |
| 6 | Npc1 | 1496 | 0.453 | 0.0906 | No |
| 7 | Gnpat | 1537 | 0.448 | 0.1110 | No |
| 8 | Cyp46a1 | 1788 | 0.424 | 0.1197 | No |
| 9 | Abca6 | 3804 | 0.271 | 0.0322 | No |
| 10 | Hao1 | 3858 | 0.268 | 0.0429 | No |
| 11 | Ar | 4008 | 0.262 | 0.0486 | No |
| 12 | Aldh9a1 | 4234 | 0.248 | 0.0497 | No |
| 13 | Klf1 | 4494 | 0.235 | 0.0485 | No |
| 14 | Scp2 | 5050 | 0.212 | 0.0312 | No |
| 15 | Acsl1 | 5193 | 0.203 | 0.0343 | No |
| 16 | Akr1d1 | 5260 | 0.199 | 0.0409 | No |
| 17 | Paox | 5327 | 0.195 | 0.0474 | No |
| 18 | Pex11a | 5383 | 0.193 | 0.0543 | No |
| 19 | Pecr | 5621 | 0.180 | 0.0514 | No |
| 20 | Abcd3 | 5960 | 0.162 | 0.0425 | No |
| 21 | Efhc1 | 6047 | 0.157 | 0.0461 | No |
| 22 | Slc23a2 | 6064 | 0.156 | 0.0531 | No |
| 23 | Ephx2 | 6083 | 0.156 | 0.0600 | No |
| 24 | Slc27a5 | 6145 | 0.154 | 0.0647 | No |
| 25 | Abcd1 | 6331 | 0.150 | 0.0629 | No |
| 26 | Abca4 | 6518 | 0.142 | 0.0607 | No |
| 27 | Pex1 | 6680 | 0.135 | 0.0594 | No |
| 28 | Fdxr | 7369 | 0.105 | 0.0301 | No |
| 29 | Acsl5 | 7493 | 0.100 | 0.0290 | No |
| 30 | Aldh1a1 | 7529 | 0.099 | 0.0322 | No |
| 31 | Aqp9 | 7740 | 0.089 | 0.0261 | No |
| 32 | Dio2 | 7825 | 0.085 | 0.0261 | No |
| 33 | Slc23a1 | 8045 | 0.076 | 0.0189 | No |
| 34 | Abca8b | 8124 | 0.073 | 0.0187 | No |
| 35 | Abcg4 | 8392 | 0.062 | 0.0083 | No |
| 36 | Nedd4 | 8572 | 0.053 | 0.0020 | No |
| 37 | Crot | 9251 | 0.028 | -0.0306 | No |
| 38 | Abcd2 | 9395 | 0.023 | -0.0367 | No |
| 39 | Pnpla8 | 9486 | 0.019 | -0.0402 | No |
| 40 | Bmp6 | 9500 | 0.019 | -0.0399 | No |
| 41 | Hsd17b11 | 9583 | 0.016 | -0.0432 | No |
| 42 | Pxmp2 | 9687 | 0.012 | -0.0478 | No |
| 43 | Idh1 | 9783 | 0.008 | -0.0522 | No |
| 44 | Abca2 | 9874 | 0.005 | -0.0564 | No |
| 45 | Gc | 10040 | 0.000 | -0.0647 | No |
| 46 | Aldh8a1 | 10044 | 0.000 | -0.0649 | No |
| 47 | Cyp27a1 | 10220 | -0.003 | -0.0735 | No |
| 48 | Hsd17b4 | 10306 | -0.006 | -0.0775 | No |
| 49 | Idh2 | 10311 | -0.006 | -0.0774 | No |
| 50 | Lonp2 | 10359 | -0.008 | -0.0794 | No |
| 51 | Pfkm | 10415 | -0.010 | -0.0816 | No |
| 52 | Pex7 | 10560 | -0.015 | -0.0881 | No |
| 53 | Fads1 | 10594 | -0.016 | -0.0889 | No |
| 54 | Isoc1 | 10957 | -0.030 | -0.1056 | No |
| 55 | Hacl1 | 11181 | -0.040 | -0.1148 | No |
| 56 | Abca3 | 11508 | -0.053 | -0.1285 | No |
| 57 | Nr1h4 | 11647 | -0.058 | -0.1325 | No |
| 58 | Phyh | 11825 | -0.066 | -0.1381 | No |
| 59 | Sult2b1 | 12004 | -0.072 | -0.1434 | No |
| 60 | Pex16 | 12036 | -0.073 | -0.1413 | No |
| 61 | Optn | 12304 | -0.083 | -0.1505 | No |
| 62 | Lck | 12912 | -0.109 | -0.1755 | No |
| 63 | Pex11g | 12980 | -0.112 | -0.1733 | No |
| 64 | Slc27a2 | 13216 | -0.119 | -0.1791 | No |
| 65 | Cyp39a1 | 13343 | -0.124 | -0.1792 | No |
| 66 | Fads2 | 13655 | -0.136 | -0.1880 | No |
| 67 | Pex26 | 13842 | -0.145 | -0.1901 | No |
| 68 | Slc22a18 | 14175 | -0.159 | -0.1988 | No |
| 69 | Retsat | 15007 | -0.199 | -0.2305 | Yes |
| 70 | Gstk1 | 15173 | -0.207 | -0.2285 | Yes |
| 71 | Amacr | 15411 | -0.215 | -0.2296 | Yes |
| 72 | Slc29a1 | 15509 | -0.219 | -0.2235 | Yes |
| 73 | Mlycd | 15714 | -0.228 | -0.2223 | Yes |
| 74 | Nr3c2 | 15931 | -0.239 | -0.2212 | Yes |
| 75 | Lipe | 15932 | -0.239 | -0.2092 | Yes |
| 76 | Pex6 | 16146 | -0.253 | -0.2072 | Yes |
| 77 | Rxra | 16238 | -0.259 | -0.1988 | Yes |
| 78 | Gclm | 16512 | -0.273 | -0.1988 | Yes |
| 79 | Pex19 | 16790 | -0.288 | -0.1983 | Yes |
| 80 | Prdx5 | 16833 | -0.290 | -0.1858 | Yes |
| 81 | Slc35b2 | 17621 | -0.337 | -0.2085 | Yes |
| 82 | Tfcp2l1 | 17627 | -0.337 | -0.1918 | Yes |
| 83 | Bcar3 | 17660 | -0.340 | -0.1764 | Yes |
| 84 | Atxn1 | 17724 | -0.345 | -0.1623 | Yes |
| 85 | Cat | 18195 | -0.385 | -0.1666 | Yes |
| 86 | Sod1 | 18474 | -0.408 | -0.1602 | Yes |
| 87 | Pex12 | 18592 | -0.419 | -0.1450 | Yes |
| 88 | Nr0b2 | 18655 | -0.423 | -0.1270 | Yes |
| 89 | Nudt12 | 19061 | -0.479 | -0.1233 | Yes |
| 90 | Hsd3b7 | 19100 | -0.485 | -0.1010 | Yes |
| 91 | Gnmt | 19284 | -0.524 | -0.0839 | Yes |
| 92 | Cyp7b1 | 19312 | -0.528 | -0.0588 | Yes |
| 93 | Pex13 | 19388 | -0.543 | -0.0354 | Yes |
| 94 | Soat2 | 19575 | -0.591 | -0.0152 | Yes |
| 95 | Abca1 | 19894 | -0.754 | 0.0066 | Yes |