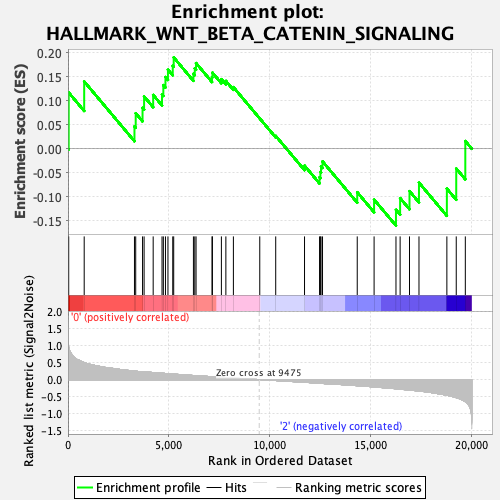
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | GMP.GMP.neu_Pheno.cls #Group1_versus_Group3.GMP.neu_Pheno.cls #Group1_versus_Group3_repos |
| Phenotype | GMP.neu_Pheno.cls#Group1_versus_Group3_repos |
| Upregulated in class | 0 |
| GeneSet | HALLMARK_WNT_BETA_CATENIN_SIGNALING |
| Enrichment Score (ES) | 0.19053021 |
| Normalized Enrichment Score (NES) | 0.61905295 |
| Nominal p-value | 0.91881186 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Tcf7 | 35 | 0.966 | 0.1172 | Yes |
| 2 | Wnt1 | 803 | 0.500 | 0.1404 | Yes |
| 3 | Notch4 | 3296 | 0.252 | 0.0467 | Yes |
| 4 | Ctnnb1 | 3358 | 0.247 | 0.0740 | Yes |
| 5 | Hdac11 | 3697 | 0.229 | 0.0853 | Yes |
| 6 | Rbpj | 3774 | 0.225 | 0.1092 | Yes |
| 7 | Adam17 | 4224 | 0.207 | 0.1122 | Yes |
| 8 | Skp2 | 4663 | 0.187 | 0.1133 | Yes |
| 9 | Nkd1 | 4728 | 0.184 | 0.1327 | Yes |
| 10 | Numb | 4833 | 0.179 | 0.1496 | Yes |
| 11 | Maml1 | 4957 | 0.174 | 0.1648 | Yes |
| 12 | Lef1 | 5195 | 0.163 | 0.1731 | Yes |
| 13 | Dvl2 | 5242 | 0.161 | 0.1905 | Yes |
| 14 | Hdac5 | 6220 | 0.120 | 0.1565 | No |
| 15 | Hdac2 | 6287 | 0.118 | 0.1677 | No |
| 16 | Cul1 | 6350 | 0.115 | 0.1787 | No |
| 17 | Jag1 | 7142 | 0.084 | 0.1495 | No |
| 18 | Notch1 | 7162 | 0.083 | 0.1588 | No |
| 19 | Ncor2 | 7607 | 0.069 | 0.1451 | No |
| 20 | Frat1 | 7829 | 0.060 | 0.1415 | No |
| 21 | Jag2 | 8202 | 0.046 | 0.1286 | No |
| 22 | Gnai1 | 9510 | 0.000 | 0.0632 | No |
| 23 | Axin1 | 10301 | -0.026 | 0.0268 | No |
| 24 | Kat2a | 11730 | -0.077 | -0.0352 | No |
| 25 | Ncstn | 12475 | -0.105 | -0.0594 | No |
| 26 | Trp53 | 12517 | -0.107 | -0.0483 | No |
| 27 | Wnt5b | 12549 | -0.108 | -0.0365 | No |
| 28 | Axin2 | 12622 | -0.112 | -0.0263 | No |
| 29 | Csnk1e | 14344 | -0.177 | -0.0907 | No |
| 30 | Ppard | 15180 | -0.215 | -0.1060 | No |
| 31 | Ptch1 | 16265 | -0.267 | -0.1273 | No |
| 32 | Wnt6 | 16471 | -0.281 | -0.1030 | No |
| 33 | Ccnd2 | 16936 | -0.307 | -0.0884 | No |
| 34 | Fzd1 | 17406 | -0.335 | -0.0707 | No |
| 35 | Psen2 | 18789 | -0.460 | -0.0832 | No |
| 36 | Myc | 19254 | -0.530 | -0.0412 | No |
| 37 | Fzd8 | 19704 | -0.648 | 0.0161 | No |