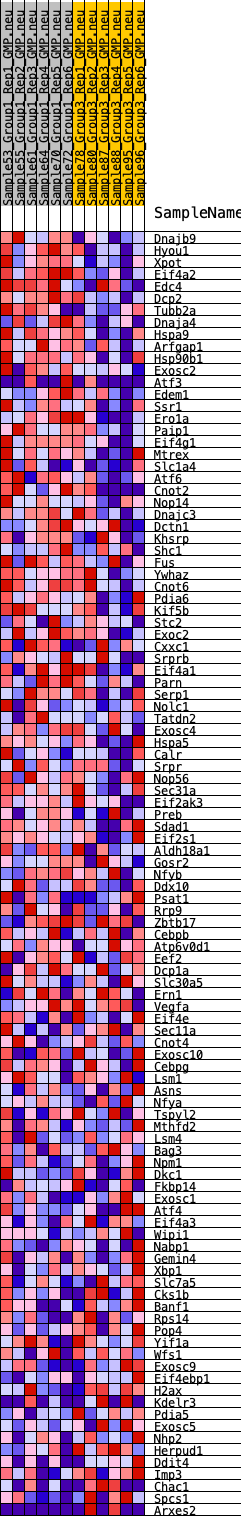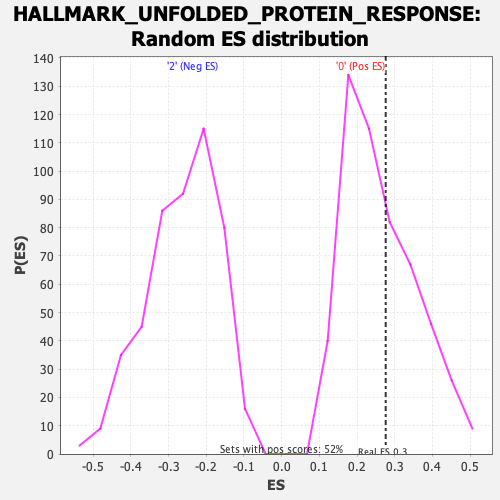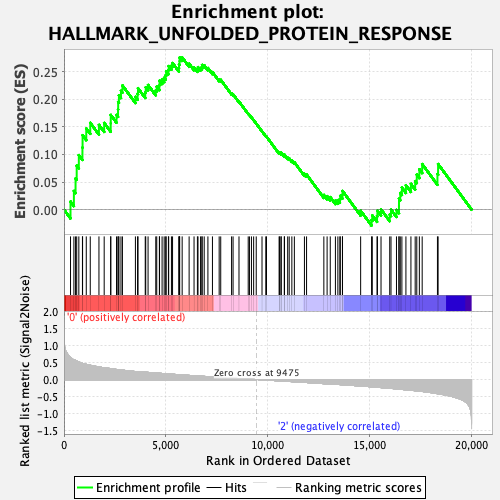
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | GMP.GMP.neu_Pheno.cls #Group1_versus_Group3.GMP.neu_Pheno.cls #Group1_versus_Group3_repos |
| Phenotype | GMP.neu_Pheno.cls#Group1_versus_Group3_repos |
| Upregulated in class | 0 |
| GeneSet | HALLMARK_UNFOLDED_PROTEIN_RESPONSE |
| Enrichment Score (ES) | 0.27597487 |
| Normalized Enrichment Score (NES) | 1.0446109 |
| Nominal p-value | 0.39499035 |
| FDR q-value | 1.0 |
| FWER p-Value | 0.972 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Dnajb9 | 320 | 0.648 | 0.0146 | Yes |
| 2 | Hyou1 | 486 | 0.583 | 0.0339 | Yes |
| 3 | Xpot | 568 | 0.564 | 0.0566 | Yes |
| 4 | Eif4a2 | 624 | 0.547 | 0.0797 | Yes |
| 5 | Edc4 | 729 | 0.515 | 0.0989 | Yes |
| 6 | Dcp2 | 906 | 0.477 | 0.1126 | Yes |
| 7 | Tubb2a | 913 | 0.477 | 0.1349 | Yes |
| 8 | Dnaja4 | 1094 | 0.450 | 0.1471 | Yes |
| 9 | Hspa9 | 1290 | 0.424 | 0.1574 | Yes |
| 10 | Arfgap1 | 1717 | 0.373 | 0.1537 | Yes |
| 11 | Hsp90b1 | 1974 | 0.346 | 0.1572 | Yes |
| 12 | Exosc2 | 2295 | 0.324 | 0.1564 | Yes |
| 13 | Atf3 | 2297 | 0.323 | 0.1717 | Yes |
| 14 | Edem1 | 2580 | 0.299 | 0.1717 | Yes |
| 15 | Ssr1 | 2655 | 0.292 | 0.1818 | Yes |
| 16 | Ero1a | 2662 | 0.292 | 0.1953 | Yes |
| 17 | Paip1 | 2699 | 0.289 | 0.2072 | Yes |
| 18 | Eif4g1 | 2799 | 0.282 | 0.2155 | Yes |
| 19 | Mtrex | 2871 | 0.278 | 0.2251 | Yes |
| 20 | Slc1a4 | 3508 | 0.237 | 0.2045 | Yes |
| 21 | Atf6 | 3615 | 0.233 | 0.2102 | Yes |
| 22 | Cnot2 | 3636 | 0.232 | 0.2202 | Yes |
| 23 | Nop14 | 3991 | 0.217 | 0.2127 | Yes |
| 24 | Dnajc3 | 4015 | 0.216 | 0.2217 | Yes |
| 25 | Dctn1 | 4130 | 0.212 | 0.2260 | Yes |
| 26 | Khsrp | 4509 | 0.194 | 0.2162 | Yes |
| 27 | Shc1 | 4556 | 0.192 | 0.2230 | Yes |
| 28 | Fus | 4686 | 0.186 | 0.2253 | Yes |
| 29 | Ywhaz | 4693 | 0.185 | 0.2338 | Yes |
| 30 | Cnot6 | 4819 | 0.180 | 0.2361 | Yes |
| 31 | Pdia6 | 4921 | 0.175 | 0.2393 | Yes |
| 32 | Kif5b | 4996 | 0.172 | 0.2437 | Yes |
| 33 | Stc2 | 5020 | 0.171 | 0.2507 | Yes |
| 34 | Exoc2 | 5130 | 0.166 | 0.2530 | Yes |
| 35 | Cxxc1 | 5143 | 0.165 | 0.2603 | Yes |
| 36 | Srprb | 5278 | 0.159 | 0.2611 | Yes |
| 37 | Eif4a1 | 5333 | 0.156 | 0.2658 | Yes |
| 38 | Parn | 5646 | 0.145 | 0.2570 | Yes |
| 39 | Serp1 | 5652 | 0.144 | 0.2635 | Yes |
| 40 | Nolc1 | 5678 | 0.143 | 0.2690 | Yes |
| 41 | Tatdn2 | 5680 | 0.143 | 0.2758 | Yes |
| 42 | Exosc4 | 5805 | 0.136 | 0.2760 | Yes |
| 43 | Hspa5 | 6147 | 0.123 | 0.2647 | No |
| 44 | Calr | 6389 | 0.113 | 0.2579 | No |
| 45 | Srpr | 6554 | 0.107 | 0.2548 | No |
| 46 | Nop56 | 6583 | 0.106 | 0.2583 | No |
| 47 | Sec31a | 6709 | 0.100 | 0.2568 | No |
| 48 | Eif2ak3 | 6776 | 0.098 | 0.2581 | No |
| 49 | Preb | 6785 | 0.098 | 0.2624 | No |
| 50 | Sdad1 | 6888 | 0.094 | 0.2617 | No |
| 51 | Eif2s1 | 7070 | 0.087 | 0.2568 | No |
| 52 | Aldh18a1 | 7293 | 0.079 | 0.2493 | No |
| 53 | Gosr2 | 7623 | 0.069 | 0.2361 | No |
| 54 | Nfyb | 7700 | 0.065 | 0.2354 | No |
| 55 | Ddx10 | 8233 | 0.045 | 0.2108 | No |
| 56 | Psat1 | 8306 | 0.042 | 0.2091 | No |
| 57 | Rrp9 | 8597 | 0.031 | 0.1961 | No |
| 58 | Zbtb17 | 9050 | 0.016 | 0.1741 | No |
| 59 | Cebpb | 9111 | 0.013 | 0.1717 | No |
| 60 | Atp6v0d1 | 9209 | 0.010 | 0.1673 | No |
| 61 | Eef2 | 9317 | 0.006 | 0.1623 | No |
| 62 | Dcp1a | 9444 | 0.001 | 0.1560 | No |
| 63 | Slc30a5 | 9728 | -0.006 | 0.1421 | No |
| 64 | Ern1 | 9914 | -0.012 | 0.1334 | No |
| 65 | Vegfa | 9948 | -0.013 | 0.1323 | No |
| 66 | Eif4e | 10568 | -0.035 | 0.1029 | No |
| 67 | Sec11a | 10591 | -0.036 | 0.1035 | No |
| 68 | Cnot4 | 10661 | -0.039 | 0.1019 | No |
| 69 | Exosc10 | 10671 | -0.039 | 0.1033 | No |
| 70 | Cebpg | 10823 | -0.045 | 0.0978 | No |
| 71 | Lsm1 | 10832 | -0.045 | 0.0995 | No |
| 72 | Asns | 10990 | -0.051 | 0.0940 | No |
| 73 | Nfya | 11064 | -0.053 | 0.0929 | No |
| 74 | Tspyl2 | 11194 | -0.057 | 0.0891 | No |
| 75 | Mthfd2 | 11317 | -0.062 | 0.0859 | No |
| 76 | Lsm4 | 11812 | -0.080 | 0.0649 | No |
| 77 | Bag3 | 11917 | -0.084 | 0.0637 | No |
| 78 | Npm1 | 12766 | -0.117 | 0.0267 | No |
| 79 | Dkc1 | 12926 | -0.121 | 0.0244 | No |
| 80 | Fkbp14 | 13081 | -0.127 | 0.0227 | No |
| 81 | Exosc1 | 13334 | -0.137 | 0.0165 | No |
| 82 | Atf4 | 13455 | -0.141 | 0.0172 | No |
| 83 | Eif4a3 | 13554 | -0.145 | 0.0191 | No |
| 84 | Wipi1 | 13573 | -0.146 | 0.0251 | No |
| 85 | Nabp1 | 13679 | -0.150 | 0.0269 | No |
| 86 | Gemin4 | 13683 | -0.150 | 0.0339 | No |
| 87 | Xbp1 | 14574 | -0.185 | -0.0021 | No |
| 88 | Slc7a5 | 15111 | -0.212 | -0.0189 | No |
| 89 | Cks1b | 15145 | -0.213 | -0.0105 | No |
| 90 | Banf1 | 15380 | -0.226 | -0.0116 | No |
| 91 | Rps14 | 15398 | -0.226 | -0.0018 | No |
| 92 | Pop4 | 15575 | -0.233 | 0.0004 | No |
| 93 | Yif1a | 16003 | -0.254 | -0.0090 | No |
| 94 | Wfs1 | 16065 | -0.257 | 0.0001 | No |
| 95 | Exosc9 | 16334 | -0.272 | -0.0005 | No |
| 96 | Eif4ebp1 | 16457 | -0.280 | 0.0067 | No |
| 97 | H2ax | 16458 | -0.280 | 0.0199 | No |
| 98 | Kdelr3 | 16521 | -0.283 | 0.0302 | No |
| 99 | Pdia5 | 16600 | -0.288 | 0.0400 | No |
| 100 | Exosc5 | 16798 | -0.299 | 0.0443 | No |
| 101 | Nhp2 | 17044 | -0.313 | 0.0468 | No |
| 102 | Herpud1 | 17253 | -0.326 | 0.0517 | No |
| 103 | Ddit4 | 17332 | -0.330 | 0.0634 | No |
| 104 | Imp3 | 17458 | -0.338 | 0.0731 | No |
| 105 | Chac1 | 17597 | -0.347 | 0.0826 | No |
| 106 | Spcs1 | 18347 | -0.413 | 0.0646 | No |
| 107 | Arxes2 | 18379 | -0.416 | 0.0827 | No |