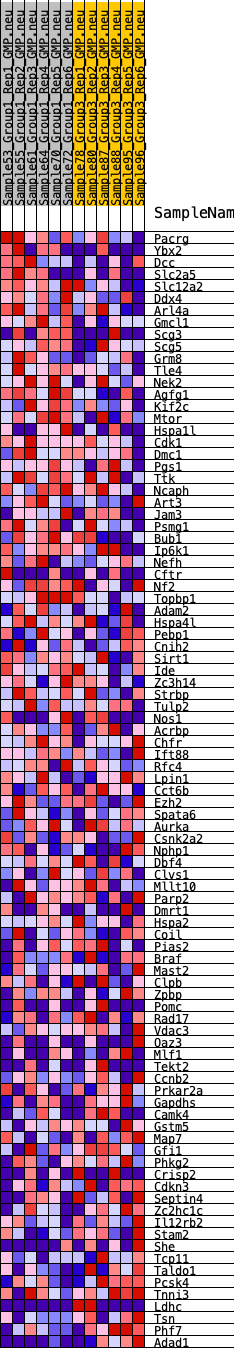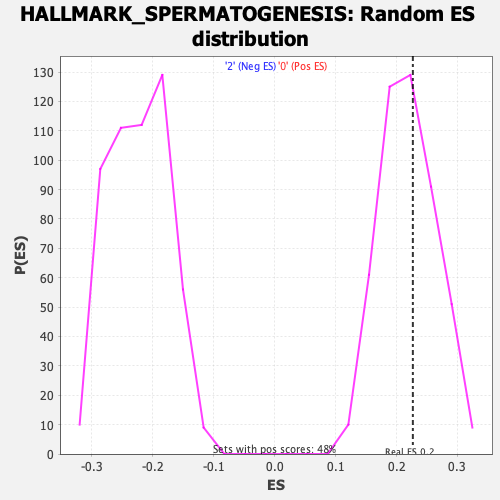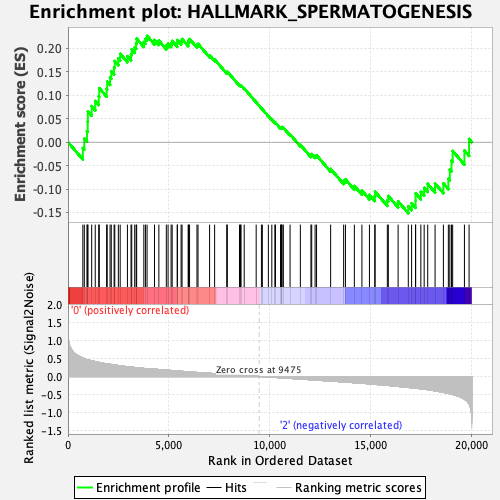
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | GMP.GMP.neu_Pheno.cls #Group1_versus_Group3.GMP.neu_Pheno.cls #Group1_versus_Group3_repos |
| Phenotype | GMP.neu_Pheno.cls#Group1_versus_Group3_repos |
| Upregulated in class | 0 |
| GeneSet | HALLMARK_SPERMATOGENESIS |
| Enrichment Score (ES) | 0.22676772 |
| Normalized Enrichment Score (NES) | 1.0329984 |
| Nominal p-value | 0.43067226 |
| FDR q-value | 0.9235754 |
| FWER p-Value | 0.98 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Pacrg | 737 | 0.514 | -0.0125 | Yes |
| 2 | Ybx2 | 809 | 0.500 | 0.0077 | Yes |
| 3 | Dcc | 940 | 0.470 | 0.0235 | Yes |
| 4 | Slc2a5 | 978 | 0.465 | 0.0438 | Yes |
| 5 | Slc12a2 | 985 | 0.464 | 0.0656 | Yes |
| 6 | Ddx4 | 1174 | 0.437 | 0.0769 | Yes |
| 7 | Arl4a | 1351 | 0.415 | 0.0878 | Yes |
| 8 | Gmcl1 | 1525 | 0.394 | 0.0979 | Yes |
| 9 | Scg3 | 1550 | 0.390 | 0.1153 | Yes |
| 10 | Scg5 | 1915 | 0.354 | 0.1138 | Yes |
| 11 | Grm8 | 1943 | 0.350 | 0.1291 | Yes |
| 12 | Tle4 | 2086 | 0.338 | 0.1381 | Yes |
| 13 | Nek2 | 2146 | 0.335 | 0.1511 | Yes |
| 14 | Agfg1 | 2286 | 0.324 | 0.1595 | Yes |
| 15 | Kif2c | 2320 | 0.320 | 0.1731 | Yes |
| 16 | Mtor | 2497 | 0.305 | 0.1788 | Yes |
| 17 | Hspa1l | 2588 | 0.298 | 0.1885 | Yes |
| 18 | Cdk1 | 2947 | 0.273 | 0.1835 | Yes |
| 19 | Dmc1 | 3126 | 0.263 | 0.1871 | Yes |
| 20 | Pgs1 | 3161 | 0.260 | 0.1977 | Yes |
| 21 | Ttk | 3308 | 0.251 | 0.2024 | Yes |
| 22 | Ncaph | 3380 | 0.245 | 0.2105 | Yes |
| 23 | Art3 | 3405 | 0.244 | 0.2209 | Yes |
| 24 | Jam3 | 3761 | 0.225 | 0.2138 | Yes |
| 25 | Psmg1 | 3840 | 0.221 | 0.2204 | Yes |
| 26 | Bub1 | 3921 | 0.218 | 0.2268 | Yes |
| 27 | Ip6k1 | 4290 | 0.204 | 0.2180 | No |
| 28 | Nefh | 4506 | 0.194 | 0.2165 | No |
| 29 | Cftr | 4876 | 0.178 | 0.2064 | No |
| 30 | Nf2 | 4963 | 0.174 | 0.2103 | No |
| 31 | Topbp1 | 5113 | 0.167 | 0.2108 | No |
| 32 | Adam2 | 5166 | 0.164 | 0.2160 | No |
| 33 | Hspa4l | 5418 | 0.154 | 0.2107 | No |
| 34 | Pebp1 | 5425 | 0.154 | 0.2178 | No |
| 35 | Cnih2 | 5609 | 0.146 | 0.2155 | No |
| 36 | Sirt1 | 5656 | 0.144 | 0.2201 | No |
| 37 | Ide | 5960 | 0.130 | 0.2110 | No |
| 38 | Zc3h14 | 5974 | 0.130 | 0.2166 | No |
| 39 | Strbp | 6028 | 0.127 | 0.2200 | No |
| 40 | Tulp2 | 6396 | 0.113 | 0.2069 | No |
| 41 | Nos1 | 6447 | 0.111 | 0.2097 | No |
| 42 | Acrbp | 7022 | 0.089 | 0.1851 | No |
| 43 | Chfr | 7271 | 0.080 | 0.1764 | No |
| 44 | Ift88 | 7872 | 0.059 | 0.1491 | No |
| 45 | Rfc4 | 7901 | 0.058 | 0.1505 | No |
| 46 | Lpin1 | 8505 | 0.034 | 0.1219 | No |
| 47 | Cct6b | 8556 | 0.033 | 0.1209 | No |
| 48 | Ezh2 | 8582 | 0.032 | 0.1212 | No |
| 49 | Spata6 | 8738 | 0.027 | 0.1147 | No |
| 50 | Aurka | 9332 | 0.006 | 0.0852 | No |
| 51 | Csnk2a2 | 9592 | -0.001 | 0.0723 | No |
| 52 | Nphp1 | 9638 | -0.003 | 0.0701 | No |
| 53 | Dbf4 | 9936 | -0.013 | 0.0559 | No |
| 54 | Clvs1 | 10108 | -0.019 | 0.0482 | No |
| 55 | Mllt10 | 10260 | -0.024 | 0.0418 | No |
| 56 | Parp2 | 10288 | -0.025 | 0.0416 | No |
| 57 | Dmrt1 | 10539 | -0.034 | 0.0307 | No |
| 58 | Hspa2 | 10585 | -0.035 | 0.0301 | No |
| 59 | Coil | 10601 | -0.036 | 0.0311 | No |
| 60 | Pias2 | 10612 | -0.036 | 0.0323 | No |
| 61 | Braf | 10680 | -0.040 | 0.0308 | No |
| 62 | Mast2 | 11013 | -0.051 | 0.0166 | No |
| 63 | Clpb | 11523 | -0.068 | -0.0057 | No |
| 64 | Zpbp | 12047 | -0.089 | -0.0277 | No |
| 65 | Pomc | 12087 | -0.090 | -0.0254 | No |
| 66 | Rad17 | 12254 | -0.096 | -0.0291 | No |
| 67 | Vdac3 | 12326 | -0.099 | -0.0280 | No |
| 68 | Oaz3 | 13025 | -0.124 | -0.0571 | No |
| 69 | Mlf1 | 13670 | -0.149 | -0.0823 | No |
| 70 | Tekt2 | 13759 | -0.153 | -0.0795 | No |
| 71 | Ccnb2 | 14201 | -0.172 | -0.0934 | No |
| 72 | Prkar2a | 14576 | -0.185 | -0.1034 | No |
| 73 | Gapdhs | 14949 | -0.203 | -0.1124 | No |
| 74 | Camk4 | 15215 | -0.217 | -0.1153 | No |
| 75 | Gstm5 | 15229 | -0.217 | -0.1056 | No |
| 76 | Map7 | 15836 | -0.246 | -0.1243 | No |
| 77 | Gfi1 | 15886 | -0.249 | -0.1149 | No |
| 78 | Phkg2 | 16371 | -0.275 | -0.1261 | No |
| 79 | Crisp2 | 16876 | -0.304 | -0.1370 | No |
| 80 | Cdkn3 | 17040 | -0.313 | -0.1303 | No |
| 81 | Septin4 | 17237 | -0.325 | -0.1247 | No |
| 82 | Zc2hc1c | 17239 | -0.325 | -0.1093 | No |
| 83 | Il12rb2 | 17499 | -0.341 | -0.1061 | No |
| 84 | Stam2 | 17663 | -0.352 | -0.0975 | No |
| 85 | She | 17839 | -0.365 | -0.0889 | No |
| 86 | Tcp11 | 18204 | -0.397 | -0.0882 | No |
| 87 | Taldo1 | 18617 | -0.440 | -0.0880 | No |
| 88 | Pcsk4 | 18863 | -0.469 | -0.0780 | No |
| 89 | Tnni3 | 18935 | -0.479 | -0.0588 | No |
| 90 | Ldhc | 19017 | -0.490 | -0.0395 | No |
| 91 | Tsn | 19076 | -0.498 | -0.0187 | No |
| 92 | Phf7 | 19660 | -0.632 | -0.0179 | No |
| 93 | Adad1 | 19890 | -0.761 | 0.0068 | No |