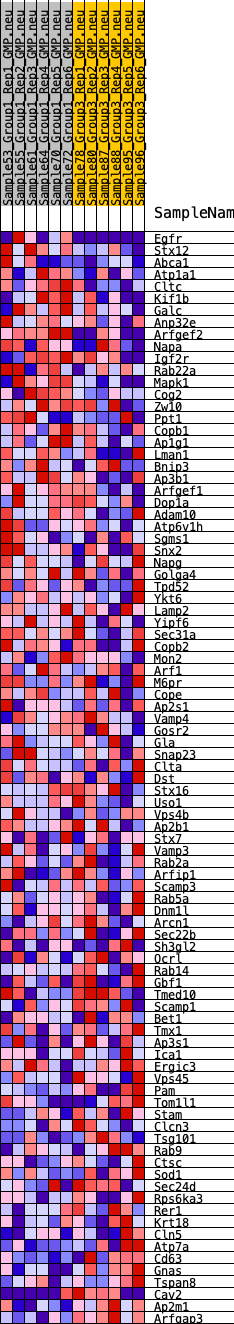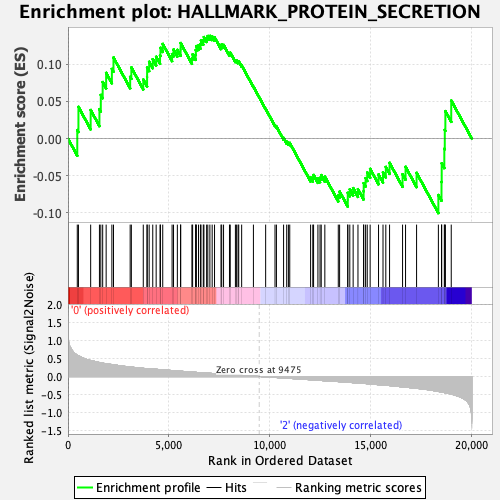
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | GMP.GMP.neu_Pheno.cls #Group1_versus_Group3.GMP.neu_Pheno.cls #Group1_versus_Group3_repos |
| Phenotype | GMP.neu_Pheno.cls#Group1_versus_Group3_repos |
| Upregulated in class | 0 |
| GeneSet | HALLMARK_PROTEIN_SECRETION |
| Enrichment Score (ES) | 0.13805515 |
| Normalized Enrichment Score (NES) | 0.66403735 |
| Nominal p-value | 0.8816794 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Egfr | 460 | 0.588 | 0.0115 | Yes |
| 2 | Stx12 | 518 | 0.577 | 0.0426 | Yes |
| 3 | Abca1 | 1124 | 0.444 | 0.0384 | Yes |
| 4 | Atp1a1 | 1559 | 0.389 | 0.0396 | Yes |
| 5 | Cltc | 1627 | 0.382 | 0.0587 | Yes |
| 6 | Kif1b | 1720 | 0.372 | 0.0760 | Yes |
| 7 | Galc | 1892 | 0.355 | 0.0884 | Yes |
| 8 | Anp32e | 2176 | 0.332 | 0.0937 | Yes |
| 9 | Arfgef2 | 2256 | 0.327 | 0.1090 | Yes |
| 10 | Napa | 3083 | 0.265 | 0.0831 | Yes |
| 11 | Igf2r | 3142 | 0.262 | 0.0956 | Yes |
| 12 | Rab22a | 3733 | 0.227 | 0.0794 | Yes |
| 13 | Mapk1 | 3922 | 0.218 | 0.0828 | Yes |
| 14 | Cog2 | 3924 | 0.218 | 0.0956 | Yes |
| 15 | Zw10 | 4025 | 0.216 | 0.1033 | Yes |
| 16 | Ppt1 | 4203 | 0.209 | 0.1067 | Yes |
| 17 | Copb1 | 4372 | 0.200 | 0.1101 | Yes |
| 18 | Ap1g1 | 4565 | 0.192 | 0.1117 | Yes |
| 19 | Lman1 | 4585 | 0.191 | 0.1220 | Yes |
| 20 | Bnip3 | 4698 | 0.185 | 0.1273 | Yes |
| 21 | Ap3b1 | 5162 | 0.165 | 0.1137 | Yes |
| 22 | Arfgef1 | 5233 | 0.161 | 0.1197 | Yes |
| 23 | Dop1a | 5423 | 0.154 | 0.1193 | Yes |
| 24 | Adam10 | 5585 | 0.147 | 0.1198 | Yes |
| 25 | Atp6v1h | 5586 | 0.147 | 0.1285 | Yes |
| 26 | Sgms1 | 6146 | 0.123 | 0.1077 | Yes |
| 27 | Snx2 | 6180 | 0.122 | 0.1132 | Yes |
| 28 | Napg | 6330 | 0.116 | 0.1125 | Yes |
| 29 | Golga4 | 6338 | 0.115 | 0.1190 | Yes |
| 30 | Tpd52 | 6366 | 0.114 | 0.1243 | Yes |
| 31 | Ykt6 | 6473 | 0.110 | 0.1255 | Yes |
| 32 | Lamp2 | 6568 | 0.106 | 0.1270 | Yes |
| 33 | Yipf6 | 6595 | 0.105 | 0.1319 | Yes |
| 34 | Sec31a | 6709 | 0.100 | 0.1322 | Yes |
| 35 | Copb2 | 6740 | 0.099 | 0.1365 | Yes |
| 36 | Mon2 | 6875 | 0.095 | 0.1353 | Yes |
| 37 | Arf1 | 6933 | 0.093 | 0.1379 | Yes |
| 38 | M6pr | 7036 | 0.089 | 0.1381 | Yes |
| 39 | Cope | 7150 | 0.084 | 0.1373 | No |
| 40 | Ap2s1 | 7267 | 0.080 | 0.1362 | No |
| 41 | Vamp4 | 7588 | 0.070 | 0.1243 | No |
| 42 | Gosr2 | 7623 | 0.069 | 0.1266 | No |
| 43 | Gla | 7714 | 0.065 | 0.1259 | No |
| 44 | Snap23 | 8008 | 0.053 | 0.1144 | No |
| 45 | Clta | 8046 | 0.052 | 0.1156 | No |
| 46 | Dst | 8302 | 0.042 | 0.1053 | No |
| 47 | Stx16 | 8351 | 0.040 | 0.1052 | No |
| 48 | Uso1 | 8432 | 0.037 | 0.1034 | No |
| 49 | Vps4b | 8467 | 0.036 | 0.1038 | No |
| 50 | Ap2b1 | 8612 | 0.031 | 0.0984 | No |
| 51 | Stx7 | 9195 | 0.011 | 0.0698 | No |
| 52 | Vamp3 | 9802 | -0.008 | 0.0399 | No |
| 53 | Rab2a | 10266 | -0.024 | 0.0181 | No |
| 54 | Arfip1 | 10337 | -0.027 | 0.0162 | No |
| 55 | Scamp3 | 10691 | -0.040 | 0.0008 | No |
| 56 | Rab5a | 10846 | -0.046 | -0.0042 | No |
| 57 | Dnm1l | 10930 | -0.049 | -0.0055 | No |
| 58 | Arcn1 | 10997 | -0.051 | -0.0058 | No |
| 59 | Sec22b | 12031 | -0.088 | -0.0525 | No |
| 60 | Sh3gl2 | 12141 | -0.092 | -0.0525 | No |
| 61 | Ocrl | 12179 | -0.093 | -0.0489 | No |
| 62 | Rab14 | 12394 | -0.102 | -0.0536 | No |
| 63 | Gbf1 | 12499 | -0.106 | -0.0525 | No |
| 64 | Tmed10 | 12558 | -0.109 | -0.0490 | No |
| 65 | Scamp1 | 12737 | -0.116 | -0.0511 | No |
| 66 | Bet1 | 13404 | -0.140 | -0.0763 | No |
| 67 | Tmx1 | 13470 | -0.142 | -0.0712 | No |
| 68 | Ap3s1 | 13874 | -0.158 | -0.0821 | No |
| 69 | Ica1 | 13875 | -0.158 | -0.0728 | No |
| 70 | Ergic3 | 13975 | -0.162 | -0.0683 | No |
| 71 | Vps45 | 14141 | -0.170 | -0.0666 | No |
| 72 | Pam | 14380 | -0.179 | -0.0680 | No |
| 73 | Tom1l1 | 14659 | -0.189 | -0.0708 | No |
| 74 | Stam | 14665 | -0.189 | -0.0599 | No |
| 75 | Clcn3 | 14759 | -0.194 | -0.0532 | No |
| 76 | Tsg101 | 14840 | -0.198 | -0.0455 | No |
| 77 | Rab9 | 14984 | -0.204 | -0.0407 | No |
| 78 | Ctsc | 15400 | -0.226 | -0.0482 | No |
| 79 | Sod1 | 15622 | -0.236 | -0.0454 | No |
| 80 | Sec24d | 15761 | -0.242 | -0.0381 | No |
| 81 | Rps6ka3 | 15947 | -0.251 | -0.0326 | No |
| 82 | Rer1 | 16592 | -0.288 | -0.0479 | No |
| 83 | Krt18 | 16740 | -0.295 | -0.0379 | No |
| 84 | Cln5 | 17287 | -0.327 | -0.0461 | No |
| 85 | Atp7a | 18368 | -0.415 | -0.0758 | No |
| 86 | Cd63 | 18523 | -0.430 | -0.0582 | No |
| 87 | Gnas | 18533 | -0.431 | -0.0333 | No |
| 88 | Tspan8 | 18663 | -0.446 | -0.0135 | No |
| 89 | Cav2 | 18686 | -0.448 | 0.0118 | No |
| 90 | Ap2m1 | 18715 | -0.452 | 0.0369 | No |
| 91 | Arfgap3 | 19006 | -0.489 | 0.0512 | No |