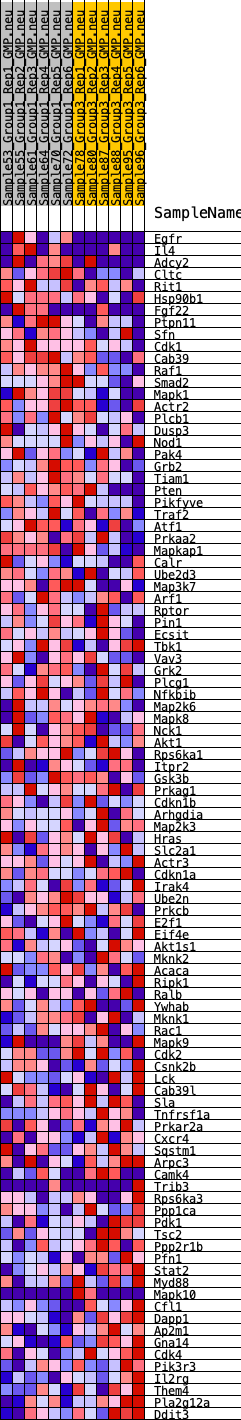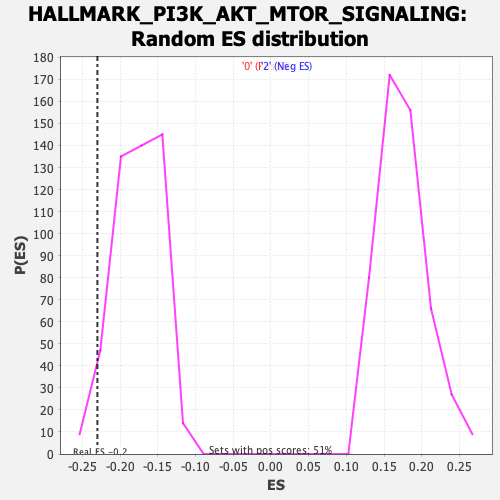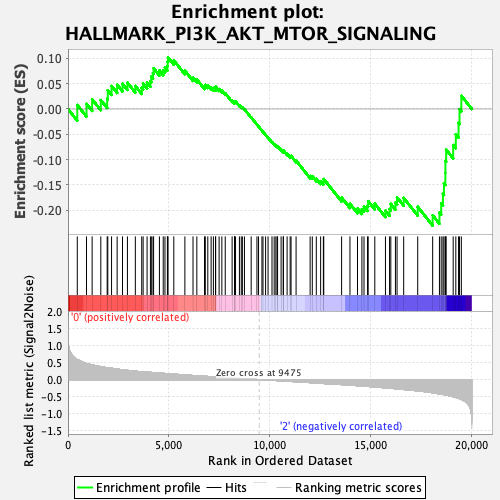
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | GMP.GMP.neu_Pheno.cls #Group1_versus_Group3.GMP.neu_Pheno.cls #Group1_versus_Group3_repos |
| Phenotype | GMP.neu_Pheno.cls#Group1_versus_Group3_repos |
| Upregulated in class | 2 |
| GeneSet | HALLMARK_PI3K_AKT_MTOR_SIGNALING |
| Enrichment Score (ES) | -0.23013633 |
| Normalized Enrichment Score (NES) | -1.3078231 |
| Nominal p-value | 0.030612245 |
| FDR q-value | 0.8846376 |
| FWER p-Value | 0.771 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Egfr | 460 | 0.588 | 0.0079 | No |
| 2 | Il4 | 917 | 0.476 | 0.0101 | No |
| 3 | Adcy2 | 1197 | 0.434 | 0.0190 | No |
| 4 | Cltc | 1627 | 0.382 | 0.0177 | No |
| 5 | Rit1 | 1942 | 0.350 | 0.0204 | No |
| 6 | Hsp90b1 | 1974 | 0.346 | 0.0371 | No |
| 7 | Fgf22 | 2160 | 0.334 | 0.0454 | No |
| 8 | Ptpn11 | 2437 | 0.310 | 0.0480 | No |
| 9 | Sfn | 2703 | 0.288 | 0.0499 | No |
| 10 | Cdk1 | 2947 | 0.273 | 0.0521 | No |
| 11 | Cab39 | 3339 | 0.248 | 0.0455 | No |
| 12 | Raf1 | 3656 | 0.231 | 0.0419 | No |
| 13 | Smad2 | 3726 | 0.227 | 0.0504 | No |
| 14 | Mapk1 | 3922 | 0.218 | 0.0521 | No |
| 15 | Actr2 | 4088 | 0.213 | 0.0551 | No |
| 16 | Plcb1 | 4132 | 0.212 | 0.0641 | No |
| 17 | Dusp3 | 4215 | 0.208 | 0.0709 | No |
| 18 | Nod1 | 4245 | 0.206 | 0.0804 | No |
| 19 | Pak4 | 4534 | 0.193 | 0.0761 | No |
| 20 | Grb2 | 4722 | 0.184 | 0.0764 | No |
| 21 | Tiam1 | 4808 | 0.181 | 0.0817 | No |
| 22 | Pten | 4930 | 0.175 | 0.0849 | No |
| 23 | Pikfyve | 4941 | 0.175 | 0.0936 | No |
| 24 | Traf2 | 4960 | 0.174 | 0.1018 | No |
| 25 | Atf1 | 5247 | 0.160 | 0.0959 | No |
| 26 | Prkaa2 | 5796 | 0.136 | 0.0756 | No |
| 27 | Mapkap1 | 6200 | 0.121 | 0.0618 | No |
| 28 | Calr | 6389 | 0.113 | 0.0583 | No |
| 29 | Ube2d3 | 6769 | 0.098 | 0.0444 | No |
| 30 | Map3k7 | 6809 | 0.097 | 0.0476 | No |
| 31 | Arf1 | 6933 | 0.093 | 0.0463 | No |
| 32 | Rptor | 7098 | 0.086 | 0.0426 | No |
| 33 | Pin1 | 7210 | 0.082 | 0.0414 | No |
| 34 | Ecsit | 7319 | 0.078 | 0.0401 | No |
| 35 | Tbk1 | 7320 | 0.078 | 0.0442 | No |
| 36 | Vav3 | 7494 | 0.073 | 0.0393 | No |
| 37 | Grk2 | 7630 | 0.069 | 0.0362 | No |
| 38 | Plcg1 | 7798 | 0.062 | 0.0311 | No |
| 39 | Nfkbib | 8139 | 0.049 | 0.0166 | No |
| 40 | Map2k6 | 8257 | 0.044 | 0.0130 | No |
| 41 | Mapk8 | 8258 | 0.044 | 0.0153 | No |
| 42 | Nck1 | 8308 | 0.042 | 0.0151 | No |
| 43 | Akt1 | 8501 | 0.035 | 0.0073 | No |
| 44 | Rps6ka1 | 8593 | 0.031 | 0.0043 | No |
| 45 | Itpr2 | 8647 | 0.029 | 0.0032 | No |
| 46 | Gsk3b | 8750 | 0.026 | -0.0005 | No |
| 47 | Prkag1 | 9084 | 0.015 | -0.0164 | No |
| 48 | Cdkn1b | 9370 | 0.004 | -0.0305 | No |
| 49 | Arhgdia | 9449 | 0.001 | -0.0344 | No |
| 50 | Map2k3 | 9617 | -0.002 | -0.0426 | No |
| 51 | Hras | 9670 | -0.004 | -0.0450 | No |
| 52 | Slc2a1 | 9799 | -0.008 | -0.0510 | No |
| 53 | Actr3 | 9921 | -0.012 | -0.0564 | No |
| 54 | Cdkn1a | 10117 | -0.019 | -0.0652 | No |
| 55 | Irak4 | 10225 | -0.023 | -0.0694 | No |
| 56 | Ube2n | 10309 | -0.026 | -0.0722 | No |
| 57 | Prkcb | 10370 | -0.028 | -0.0737 | No |
| 58 | E2f1 | 10393 | -0.029 | -0.0733 | No |
| 59 | Eif4e | 10568 | -0.035 | -0.0802 | No |
| 60 | Akt1s1 | 10674 | -0.039 | -0.0834 | No |
| 61 | Mknk2 | 10682 | -0.040 | -0.0816 | No |
| 62 | Acaca | 10866 | -0.046 | -0.0884 | No |
| 63 | Ripk1 | 11017 | -0.052 | -0.0932 | No |
| 64 | Ralb | 11062 | -0.053 | -0.0926 | No |
| 65 | Ywhab | 11311 | -0.061 | -0.1018 | No |
| 66 | Mknk1 | 12008 | -0.087 | -0.1322 | No |
| 67 | Rac1 | 12115 | -0.091 | -0.1327 | No |
| 68 | Mapk9 | 12314 | -0.098 | -0.1374 | No |
| 69 | Cdk2 | 12528 | -0.108 | -0.1424 | No |
| 70 | Csnk2b | 12664 | -0.113 | -0.1432 | No |
| 71 | Lck | 12684 | -0.114 | -0.1382 | No |
| 72 | Cab39l | 13569 | -0.145 | -0.1749 | No |
| 73 | Sla | 13983 | -0.162 | -0.1870 | No |
| 74 | Tnfrsf1a | 14357 | -0.178 | -0.1964 | No |
| 75 | Prkar2a | 14576 | -0.185 | -0.1975 | No |
| 76 | Cxcr4 | 14680 | -0.190 | -0.1927 | No |
| 77 | Sqstm1 | 14852 | -0.198 | -0.1908 | No |
| 78 | Arpc3 | 14887 | -0.200 | -0.1819 | No |
| 79 | Camk4 | 15215 | -0.217 | -0.1869 | No |
| 80 | Trib3 | 15743 | -0.242 | -0.2006 | No |
| 81 | Rps6ka3 | 15947 | -0.251 | -0.1976 | No |
| 82 | Ppp1ca | 16009 | -0.254 | -0.1872 | No |
| 83 | Pdk1 | 16238 | -0.266 | -0.1847 | No |
| 84 | Tsc2 | 16319 | -0.271 | -0.1744 | No |
| 85 | Ppp2r1b | 16647 | -0.291 | -0.1755 | No |
| 86 | Pfn1 | 17343 | -0.330 | -0.1930 | No |
| 87 | Stat2 | 18085 | -0.387 | -0.2097 | Yes |
| 88 | Myd88 | 18421 | -0.420 | -0.2044 | Yes |
| 89 | Mapk10 | 18507 | -0.429 | -0.1860 | Yes |
| 90 | Cfl1 | 18594 | -0.436 | -0.1673 | Yes |
| 91 | Dapp1 | 18645 | -0.443 | -0.1465 | Yes |
| 92 | Ap2m1 | 18715 | -0.452 | -0.1262 | Yes |
| 93 | Gna14 | 18717 | -0.452 | -0.1024 | Yes |
| 94 | Cdk4 | 18754 | -0.455 | -0.0802 | Yes |
| 95 | Pik3r3 | 19102 | -0.502 | -0.0711 | Yes |
| 96 | Il2rg | 19233 | -0.526 | -0.0499 | Yes |
| 97 | Them4 | 19368 | -0.552 | -0.0275 | Yes |
| 98 | Pla2g12a | 19417 | -0.562 | -0.0003 | Yes |
| 99 | Ddit3 | 19507 | -0.585 | 0.0260 | Yes |