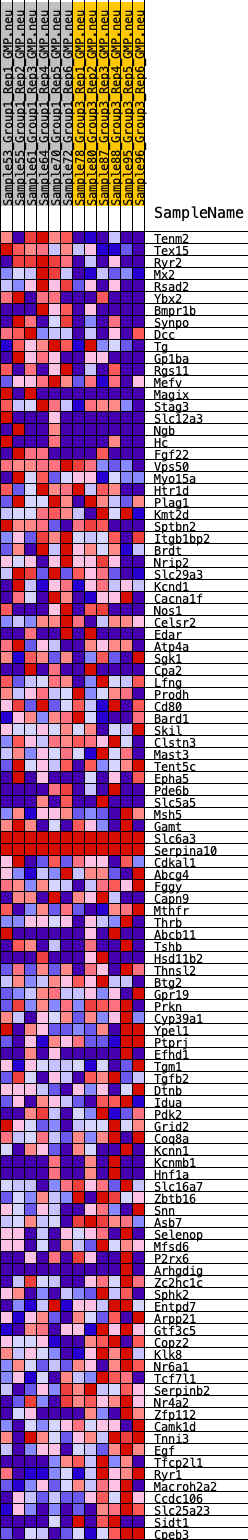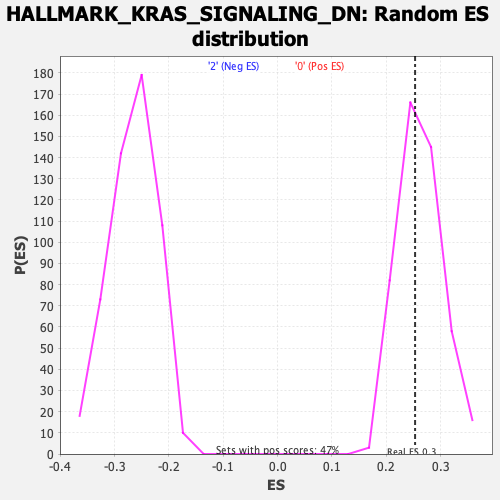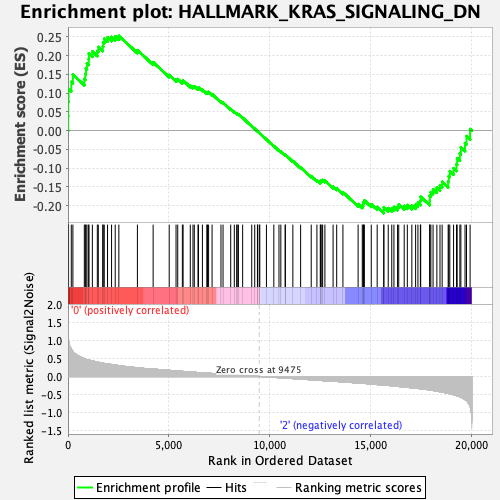
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | GMP.GMP.neu_Pheno.cls #Group1_versus_Group3.GMP.neu_Pheno.cls #Group1_versus_Group3_repos |
| Phenotype | GMP.neu_Pheno.cls#Group1_versus_Group3_repos |
| Upregulated in class | 0 |
| GeneSet | HALLMARK_KRAS_SIGNALING_DN |
| Enrichment Score (ES) | 0.2529442 |
| Normalized Enrichment Score (NES) | 0.9669188 |
| Nominal p-value | 0.56808513 |
| FDR q-value | 1.0 |
| FWER p-Value | 0.993 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Tenm2 | 9 | 1.179 | 0.0390 | Yes |
| 2 | Tex15 | 13 | 1.164 | 0.0778 | Yes |
| 3 | Ryr2 | 28 | 1.026 | 0.1114 | Yes |
| 4 | Mx2 | 163 | 0.765 | 0.1302 | Yes |
| 5 | Rsad2 | 242 | 0.695 | 0.1495 | Yes |
| 6 | Ybx2 | 809 | 0.500 | 0.1378 | Yes |
| 7 | Bmpr1b | 859 | 0.488 | 0.1517 | Yes |
| 8 | Synpo | 889 | 0.481 | 0.1663 | Yes |
| 9 | Dcc | 940 | 0.470 | 0.1795 | Yes |
| 10 | Tg | 1028 | 0.458 | 0.1905 | Yes |
| 11 | Gp1ba | 1035 | 0.457 | 0.2055 | Yes |
| 12 | Rgs11 | 1208 | 0.432 | 0.2113 | Yes |
| 13 | Mefv | 1454 | 0.401 | 0.2124 | Yes |
| 14 | Magix | 1503 | 0.395 | 0.2232 | Yes |
| 15 | Stag3 | 1722 | 0.372 | 0.2247 | Yes |
| 16 | Slc12a3 | 1745 | 0.370 | 0.2359 | Yes |
| 17 | Ngb | 1797 | 0.362 | 0.2455 | Yes |
| 18 | Hc | 1960 | 0.348 | 0.2490 | Yes |
| 19 | Fgf22 | 2160 | 0.334 | 0.2502 | Yes |
| 20 | Vps50 | 2341 | 0.319 | 0.2518 | Yes |
| 21 | Myo15a | 2522 | 0.304 | 0.2529 | Yes |
| 22 | Htr1d | 3442 | 0.241 | 0.2149 | No |
| 23 | Plag1 | 4222 | 0.208 | 0.1827 | No |
| 24 | Kmt2d | 5019 | 0.171 | 0.1484 | No |
| 25 | Sptbn2 | 5358 | 0.155 | 0.1367 | No |
| 26 | Itgb1bp2 | 5442 | 0.153 | 0.1376 | No |
| 27 | Brdt | 5671 | 0.143 | 0.1310 | No |
| 28 | Nrip2 | 5708 | 0.141 | 0.1339 | No |
| 29 | Slc29a3 | 6064 | 0.126 | 0.1203 | No |
| 30 | Kcnd1 | 6198 | 0.121 | 0.1177 | No |
| 31 | Cacna1f | 6263 | 0.118 | 0.1184 | No |
| 32 | Nos1 | 6447 | 0.111 | 0.1129 | No |
| 33 | Celsr2 | 6472 | 0.110 | 0.1154 | No |
| 34 | Edar | 6666 | 0.102 | 0.1091 | No |
| 35 | Atp4a | 6884 | 0.094 | 0.1014 | No |
| 36 | Sgk1 | 6919 | 0.093 | 0.1028 | No |
| 37 | Cpa2 | 6972 | 0.091 | 0.1033 | No |
| 38 | Lfng | 7145 | 0.084 | 0.0974 | No |
| 39 | Prodh | 7586 | 0.070 | 0.0777 | No |
| 40 | Cd80 | 7689 | 0.066 | 0.0748 | No |
| 41 | Bard1 | 8069 | 0.052 | 0.0575 | No |
| 42 | Skil | 8248 | 0.044 | 0.0500 | No |
| 43 | Clstn3 | 8366 | 0.039 | 0.0454 | No |
| 44 | Mast3 | 8390 | 0.038 | 0.0456 | No |
| 45 | Tent5c | 8452 | 0.036 | 0.0437 | No |
| 46 | Epha5 | 8661 | 0.029 | 0.0342 | No |
| 47 | Pde6b | 9112 | 0.013 | 0.0121 | No |
| 48 | Slc5a5 | 9260 | 0.008 | 0.0050 | No |
| 49 | Msh5 | 9395 | 0.003 | -0.0016 | No |
| 50 | Gamt | 9401 | 0.003 | -0.0018 | No |
| 51 | Slc6a3 | 9484 | 0.000 | -0.0059 | No |
| 52 | Serpina10 | 9511 | 0.000 | -0.0072 | No |
| 53 | Cdkal1 | 9841 | -0.010 | -0.0234 | No |
| 54 | Abcg4 | 10207 | -0.022 | -0.0410 | No |
| 55 | Fggy | 10463 | -0.031 | -0.0527 | No |
| 56 | Capn9 | 10555 | -0.035 | -0.0561 | No |
| 57 | Mthfr | 10770 | -0.043 | -0.0654 | No |
| 58 | Thrb | 10779 | -0.043 | -0.0644 | No |
| 59 | Abcb11 | 11151 | -0.056 | -0.0812 | No |
| 60 | Tshb | 11536 | -0.069 | -0.0981 | No |
| 61 | Hsd11b2 | 12062 | -0.089 | -0.1215 | No |
| 62 | Thnsl2 | 12344 | -0.100 | -0.1323 | No |
| 63 | Btg2 | 12511 | -0.107 | -0.1370 | No |
| 64 | Gpr19 | 12526 | -0.108 | -0.1341 | No |
| 65 | Prkn | 12586 | -0.110 | -0.1334 | No |
| 66 | Cyp39a1 | 12628 | -0.112 | -0.1317 | No |
| 67 | Ypel1 | 12740 | -0.116 | -0.1334 | No |
| 68 | Ptprj | 13146 | -0.129 | -0.1494 | No |
| 69 | Efhd1 | 13325 | -0.136 | -0.1538 | No |
| 70 | Tgm1 | 13634 | -0.148 | -0.1643 | No |
| 71 | Tgfb2 | 14387 | -0.179 | -0.1961 | No |
| 72 | Dtnb | 14594 | -0.186 | -0.2002 | No |
| 73 | Idua | 14634 | -0.188 | -0.1959 | No |
| 74 | Pdk2 | 14655 | -0.189 | -0.1906 | No |
| 75 | Grid2 | 14698 | -0.191 | -0.1863 | No |
| 76 | Coq8a | 15039 | -0.207 | -0.1964 | No |
| 77 | Kcnn1 | 15333 | -0.223 | -0.2037 | No |
| 78 | Kcnmb1 | 15661 | -0.237 | -0.2122 | No |
| 79 | Hnf1a | 15663 | -0.237 | -0.2043 | No |
| 80 | Slc16a7 | 15881 | -0.249 | -0.2068 | No |
| 81 | Zbtb16 | 16056 | -0.256 | -0.2070 | No |
| 82 | Snn | 16164 | -0.262 | -0.2036 | No |
| 83 | Asb7 | 16343 | -0.272 | -0.2034 | No |
| 84 | Selenop | 16402 | -0.277 | -0.1971 | No |
| 85 | Mfsd6 | 16673 | -0.292 | -0.2009 | No |
| 86 | P2rx6 | 16828 | -0.301 | -0.1985 | No |
| 87 | Arhgdig | 17052 | -0.313 | -0.1992 | No |
| 88 | Zc2hc1c | 17239 | -0.325 | -0.1977 | No |
| 89 | Sphk2 | 17354 | -0.331 | -0.1924 | No |
| 90 | Entpd7 | 17475 | -0.339 | -0.1871 | No |
| 91 | Arpp21 | 17491 | -0.340 | -0.1765 | No |
| 92 | Gtf3c5 | 17940 | -0.374 | -0.1864 | No |
| 93 | Copz2 | 17942 | -0.375 | -0.1740 | No |
| 94 | Klk8 | 17995 | -0.378 | -0.1639 | No |
| 95 | Nr6a1 | 18104 | -0.388 | -0.1563 | No |
| 96 | Tcf7l1 | 18293 | -0.407 | -0.1522 | No |
| 97 | Serpinb2 | 18448 | -0.424 | -0.1457 | No |
| 98 | Nr4a2 | 18555 | -0.433 | -0.1366 | No |
| 99 | Zfp112 | 18853 | -0.468 | -0.1358 | No |
| 100 | Camk1d | 18882 | -0.472 | -0.1214 | No |
| 101 | Tnni3 | 18935 | -0.479 | -0.1080 | No |
| 102 | Egf | 19121 | -0.506 | -0.1004 | No |
| 103 | Tfcp2l1 | 19266 | -0.532 | -0.0899 | No |
| 104 | Ryr1 | 19299 | -0.539 | -0.0734 | No |
| 105 | Macroh2a2 | 19434 | -0.565 | -0.0612 | No |
| 106 | Ccdc106 | 19483 | -0.579 | -0.0443 | No |
| 107 | Slc25a23 | 19690 | -0.642 | -0.0332 | No |
| 108 | Sidt1 | 19763 | -0.667 | -0.0145 | No |
| 109 | Cpeb3 | 19941 | -0.827 | 0.0043 | No |