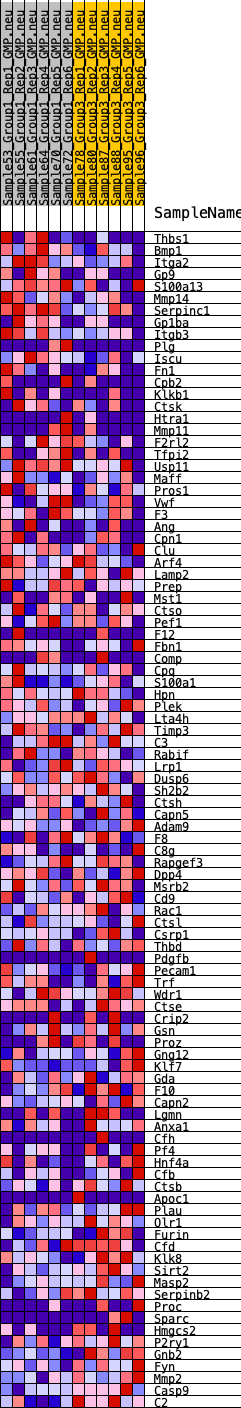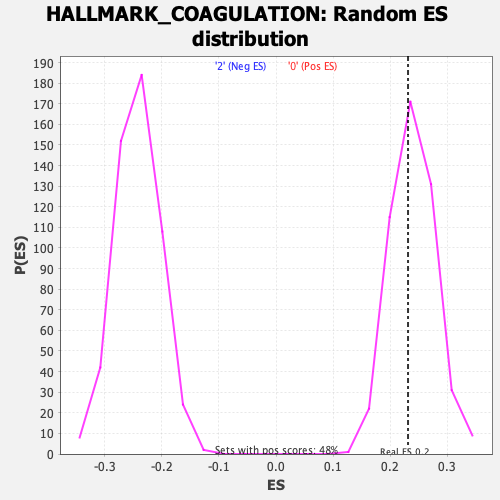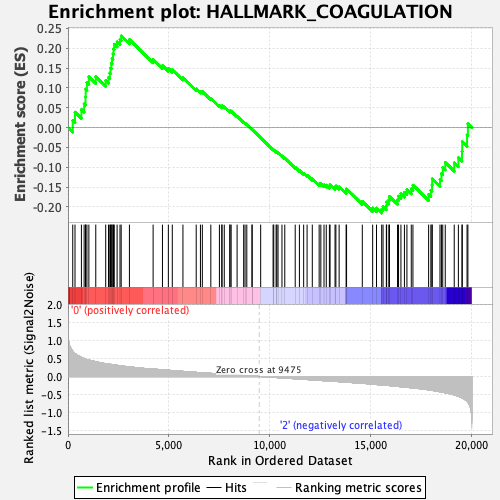
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | GMP.GMP.neu_Pheno.cls #Group1_versus_Group3.GMP.neu_Pheno.cls #Group1_versus_Group3_repos |
| Phenotype | GMP.neu_Pheno.cls#Group1_versus_Group3_repos |
| Upregulated in class | 0 |
| GeneSet | HALLMARK_COAGULATION |
| Enrichment Score (ES) | 0.23109548 |
| Normalized Enrichment Score (NES) | 0.96364385 |
| Nominal p-value | 0.57708335 |
| FDR q-value | 0.89670616 |
| FWER p-Value | 0.993 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Thbs1 | 235 | 0.704 | 0.0179 | Yes |
| 2 | Bmp1 | 347 | 0.634 | 0.0390 | Yes |
| 3 | Itga2 | 665 | 0.532 | 0.0455 | Yes |
| 4 | Gp9 | 800 | 0.501 | 0.0599 | Yes |
| 5 | S100a13 | 869 | 0.485 | 0.0769 | Yes |
| 6 | Mmp14 | 876 | 0.483 | 0.0969 | Yes |
| 7 | Serpinc1 | 938 | 0.471 | 0.1137 | Yes |
| 8 | Gp1ba | 1035 | 0.457 | 0.1281 | Yes |
| 9 | Itgb3 | 1377 | 0.411 | 0.1284 | Yes |
| 10 | Plg | 1870 | 0.356 | 0.1187 | Yes |
| 11 | Iscu | 2009 | 0.344 | 0.1262 | Yes |
| 12 | Fn1 | 2073 | 0.339 | 0.1374 | Yes |
| 13 | Cpb2 | 2108 | 0.338 | 0.1499 | Yes |
| 14 | Klkb1 | 2148 | 0.335 | 0.1620 | Yes |
| 15 | Ctsk | 2186 | 0.331 | 0.1741 | Yes |
| 16 | Htra1 | 2231 | 0.329 | 0.1858 | Yes |
| 17 | Mmp11 | 2257 | 0.327 | 0.1983 | Yes |
| 18 | F2rl2 | 2294 | 0.324 | 0.2101 | Yes |
| 19 | Tfpi2 | 2435 | 0.311 | 0.2162 | Yes |
| 20 | Usp11 | 2579 | 0.299 | 0.2216 | Yes |
| 21 | Maff | 2637 | 0.294 | 0.2311 | Yes |
| 22 | Pros1 | 3046 | 0.267 | 0.2219 | No |
| 23 | Vwf | 4221 | 0.208 | 0.1717 | No |
| 24 | F3 | 4684 | 0.186 | 0.1564 | No |
| 25 | Ang | 4977 | 0.173 | 0.1490 | No |
| 26 | Cpn1 | 5172 | 0.164 | 0.1462 | No |
| 27 | Clu | 5700 | 0.142 | 0.1257 | No |
| 28 | Arf4 | 6358 | 0.114 | 0.0975 | No |
| 29 | Lamp2 | 6568 | 0.106 | 0.0915 | No |
| 30 | Prep | 6662 | 0.103 | 0.0912 | No |
| 31 | Mst1 | 7083 | 0.087 | 0.0738 | No |
| 32 | Ctso | 7509 | 0.072 | 0.0555 | No |
| 33 | Pef1 | 7627 | 0.069 | 0.0525 | No |
| 34 | F12 | 7638 | 0.068 | 0.0549 | No |
| 35 | Fbn1 | 7750 | 0.063 | 0.0520 | No |
| 36 | Comp | 8024 | 0.053 | 0.0405 | No |
| 37 | Cpq | 8035 | 0.053 | 0.0423 | No |
| 38 | S100a1 | 8091 | 0.051 | 0.0416 | No |
| 39 | Hpn | 8386 | 0.039 | 0.0285 | No |
| 40 | Plek | 8712 | 0.028 | 0.0134 | No |
| 41 | Lta4h | 8780 | 0.025 | 0.0111 | No |
| 42 | Timp3 | 8867 | 0.022 | 0.0077 | No |
| 43 | C3 | 9128 | 0.013 | -0.0048 | No |
| 44 | Rabif | 9129 | 0.013 | -0.0043 | No |
| 45 | Lrp1 | 9555 | -0.000 | -0.0256 | No |
| 46 | Dusp6 | 10171 | -0.021 | -0.0556 | No |
| 47 | Sh2b2 | 10204 | -0.022 | -0.0563 | No |
| 48 | Ctsh | 10320 | -0.026 | -0.0609 | No |
| 49 | Capn5 | 10357 | -0.027 | -0.0616 | No |
| 50 | Adam9 | 10416 | -0.030 | -0.0632 | No |
| 51 | F8 | 10611 | -0.036 | -0.0714 | No |
| 52 | C8g | 10750 | -0.042 | -0.0766 | No |
| 53 | Rapgef3 | 11268 | -0.060 | -0.1000 | No |
| 54 | Dpp4 | 11476 | -0.067 | -0.1076 | No |
| 55 | Msrb2 | 11687 | -0.075 | -0.1150 | No |
| 56 | Cd9 | 11857 | -0.082 | -0.1200 | No |
| 57 | Rac1 | 12115 | -0.091 | -0.1291 | No |
| 58 | Ctsl | 12455 | -0.105 | -0.1417 | No |
| 59 | Csrp1 | 12539 | -0.108 | -0.1413 | No |
| 60 | Thbd | 12696 | -0.115 | -0.1443 | No |
| 61 | Pdgfb | 12808 | -0.119 | -0.1448 | No |
| 62 | Pecam1 | 12964 | -0.122 | -0.1475 | No |
| 63 | Trf | 12996 | -0.123 | -0.1438 | No |
| 64 | Wdr1 | 13243 | -0.133 | -0.1506 | No |
| 65 | Ctse | 13285 | -0.135 | -0.1470 | No |
| 66 | Crip2 | 13446 | -0.141 | -0.1490 | No |
| 67 | Gsn | 13792 | -0.154 | -0.1599 | No |
| 68 | Proz | 13815 | -0.155 | -0.1544 | No |
| 69 | Gng12 | 14595 | -0.186 | -0.1857 | No |
| 70 | Klf7 | 15109 | -0.211 | -0.2025 | No |
| 71 | Gda | 15300 | -0.222 | -0.2027 | No |
| 72 | F10 | 15556 | -0.232 | -0.2057 | No |
| 73 | Capn2 | 15624 | -0.236 | -0.1991 | No |
| 74 | Lgmn | 15788 | -0.244 | -0.1971 | No |
| 75 | Anxa1 | 15802 | -0.245 | -0.1874 | No |
| 76 | Cfh | 15912 | -0.250 | -0.1823 | No |
| 77 | Pf4 | 15942 | -0.251 | -0.1732 | No |
| 78 | Hnf4a | 16342 | -0.272 | -0.1818 | No |
| 79 | Cfb | 16392 | -0.276 | -0.1726 | No |
| 80 | Ctsb | 16511 | -0.283 | -0.1666 | No |
| 81 | Apoc1 | 16684 | -0.292 | -0.1629 | No |
| 82 | Plau | 16808 | -0.300 | -0.1565 | No |
| 83 | Olr1 | 17024 | -0.311 | -0.1541 | No |
| 84 | Furin | 17106 | -0.317 | -0.1449 | No |
| 85 | Cfd | 17884 | -0.370 | -0.1683 | No |
| 86 | Klk8 | 17995 | -0.378 | -0.1579 | No |
| 87 | Sirt2 | 18051 | -0.383 | -0.1445 | No |
| 88 | Masp2 | 18062 | -0.384 | -0.1288 | No |
| 89 | Serpinb2 | 18448 | -0.424 | -0.1303 | No |
| 90 | Proc | 18520 | -0.430 | -0.1157 | No |
| 91 | Sparc | 18585 | -0.435 | -0.1006 | No |
| 92 | Hmgcs2 | 18709 | -0.451 | -0.0878 | No |
| 93 | P2ry1 | 19154 | -0.510 | -0.0886 | No |
| 94 | Gnb2 | 19360 | -0.550 | -0.0757 | No |
| 95 | Fyn | 19539 | -0.593 | -0.0596 | No |
| 96 | Mmp2 | 19552 | -0.594 | -0.0352 | No |
| 97 | Casp9 | 19790 | -0.683 | -0.0183 | No |
| 98 | C2 | 19832 | -0.715 | 0.0097 | No |