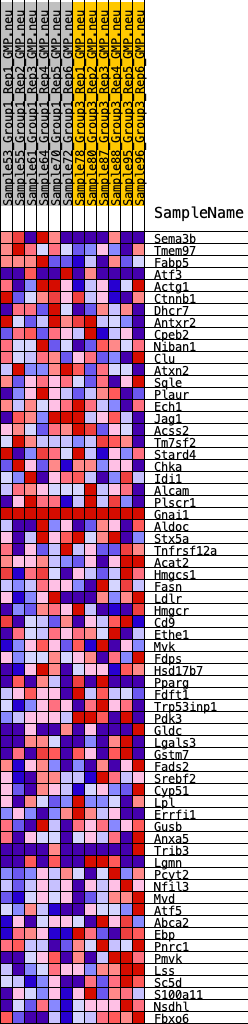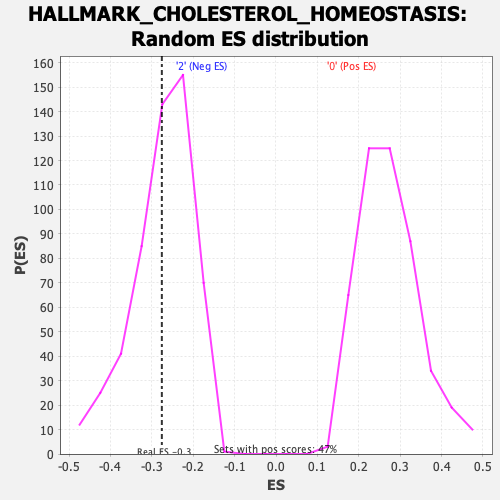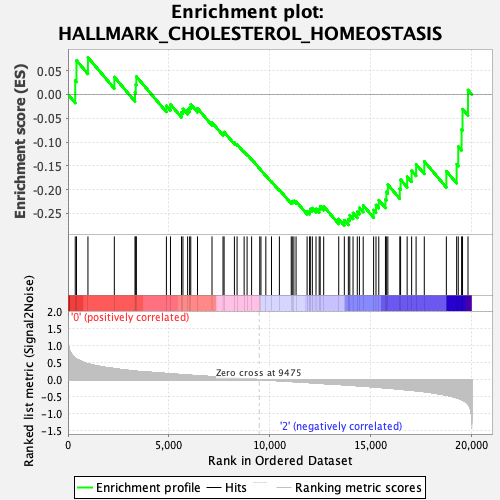
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | GMP.GMP.neu_Pheno.cls #Group1_versus_Group3.GMP.neu_Pheno.cls #Group1_versus_Group3_repos |
| Phenotype | GMP.neu_Pheno.cls#Group1_versus_Group3_repos |
| Upregulated in class | 2 |
| GeneSet | HALLMARK_CHOLESTEROL_HOMEOSTASIS |
| Enrichment Score (ES) | -0.27579057 |
| Normalized Enrichment Score (NES) | -1.0089307 |
| Nominal p-value | 0.41917294 |
| FDR q-value | 0.7587868 |
| FWER p-Value | 0.99 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Sema3b | 359 | 0.631 | 0.0294 | No |
| 2 | Tmem97 | 420 | 0.602 | 0.0716 | No |
| 3 | Fabp5 | 989 | 0.464 | 0.0780 | No |
| 4 | Atf3 | 2297 | 0.323 | 0.0368 | No |
| 5 | Actg1 | 3323 | 0.250 | 0.0041 | No |
| 6 | Ctnnb1 | 3358 | 0.247 | 0.0209 | No |
| 7 | Dhcr7 | 3390 | 0.245 | 0.0378 | No |
| 8 | Antxr2 | 4880 | 0.177 | -0.0235 | No |
| 9 | Cpeb2 | 5082 | 0.168 | -0.0209 | No |
| 10 | Niban1 | 5629 | 0.145 | -0.0374 | No |
| 11 | Clu | 5700 | 0.142 | -0.0303 | No |
| 12 | Atxn2 | 5932 | 0.131 | -0.0320 | No |
| 13 | Sqle | 6032 | 0.127 | -0.0274 | No |
| 14 | Plaur | 6088 | 0.125 | -0.0208 | No |
| 15 | Ech1 | 6422 | 0.112 | -0.0291 | No |
| 16 | Jag1 | 7142 | 0.084 | -0.0587 | No |
| 17 | Acss2 | 7688 | 0.066 | -0.0811 | No |
| 18 | Tm7sf2 | 7741 | 0.064 | -0.0789 | No |
| 19 | Stard4 | 8252 | 0.044 | -0.1012 | No |
| 20 | Chka | 8385 | 0.039 | -0.1049 | No |
| 21 | Idi1 | 8735 | 0.027 | -0.1204 | No |
| 22 | Alcam | 8881 | 0.021 | -0.1260 | No |
| 23 | Plscr1 | 9105 | 0.014 | -0.1361 | No |
| 24 | Gnai1 | 9510 | 0.000 | -0.1564 | No |
| 25 | Aldoc | 9568 | -0.001 | -0.1592 | No |
| 26 | Stx5a | 9811 | -0.009 | -0.1707 | No |
| 27 | Tnfrsf12a | 10092 | -0.018 | -0.1833 | No |
| 28 | Acat2 | 10483 | -0.032 | -0.2005 | No |
| 29 | Hmgcs1 | 11071 | -0.054 | -0.2259 | No |
| 30 | Fasn | 11122 | -0.055 | -0.2242 | No |
| 31 | Ldlr | 11202 | -0.058 | -0.2239 | No |
| 32 | Hmgcr | 11310 | -0.061 | -0.2246 | No |
| 33 | Cd9 | 11857 | -0.082 | -0.2459 | No |
| 34 | Ethe1 | 11981 | -0.086 | -0.2456 | No |
| 35 | Mvk | 12017 | -0.088 | -0.2407 | No |
| 36 | Fdps | 12117 | -0.091 | -0.2389 | No |
| 37 | Hsd17b7 | 12299 | -0.098 | -0.2406 | No |
| 38 | Pparg | 12457 | -0.105 | -0.2406 | No |
| 39 | Fdft1 | 12506 | -0.107 | -0.2350 | No |
| 40 | Trp53inp1 | 12680 | -0.114 | -0.2351 | No |
| 41 | Pdk3 | 13426 | -0.140 | -0.2619 | No |
| 42 | Gldc | 13704 | -0.150 | -0.2645 | Yes |
| 43 | Lgals3 | 13903 | -0.159 | -0.2625 | Yes |
| 44 | Gstm7 | 13974 | -0.162 | -0.2539 | Yes |
| 45 | Fads2 | 14136 | -0.169 | -0.2492 | Yes |
| 46 | Srebf2 | 14346 | -0.177 | -0.2464 | Yes |
| 47 | Cyp51 | 14452 | -0.181 | -0.2380 | Yes |
| 48 | Lpl | 14640 | -0.188 | -0.2333 | Yes |
| 49 | Errfi1 | 15156 | -0.214 | -0.2431 | Yes |
| 50 | Gusb | 15274 | -0.220 | -0.2324 | Yes |
| 51 | Anxa5 | 15412 | -0.226 | -0.2223 | Yes |
| 52 | Trib3 | 15743 | -0.242 | -0.2207 | Yes |
| 53 | Lgmn | 15788 | -0.244 | -0.2046 | Yes |
| 54 | Pcyt2 | 15862 | -0.248 | -0.1897 | Yes |
| 55 | Nfil3 | 16455 | -0.280 | -0.1983 | Yes |
| 56 | Mvd | 16494 | -0.282 | -0.1790 | Yes |
| 57 | Atf5 | 16819 | -0.301 | -0.1727 | Yes |
| 58 | Abca2 | 17042 | -0.313 | -0.1604 | Yes |
| 59 | Ebp | 17263 | -0.326 | -0.1469 | Yes |
| 60 | Pnrc1 | 17669 | -0.352 | -0.1407 | Yes |
| 61 | Pmvk | 18763 | -0.456 | -0.1613 | Yes |
| 62 | Lss | 19274 | -0.534 | -0.1467 | Yes |
| 63 | Sc5d | 19354 | -0.550 | -0.1094 | Yes |
| 64 | S100a11 | 19519 | -0.587 | -0.0736 | Yes |
| 65 | Nsdhl | 19566 | -0.600 | -0.0308 | Yes |
| 66 | Fbxo6 | 19837 | -0.717 | 0.0095 | Yes |