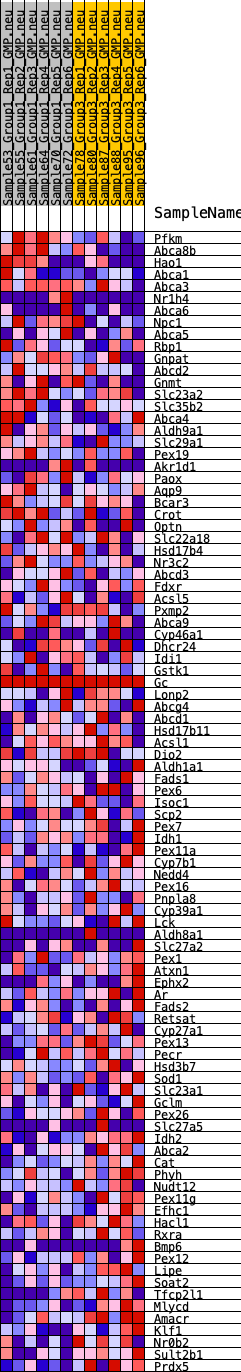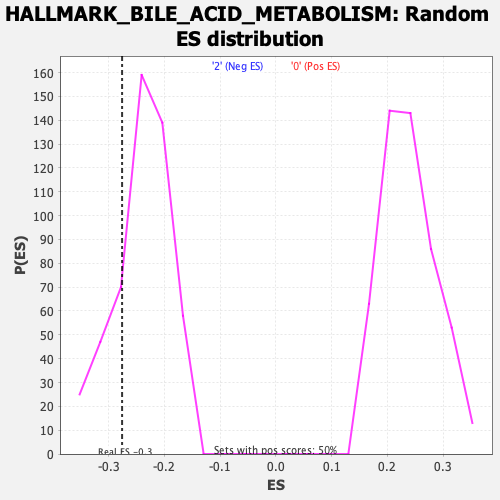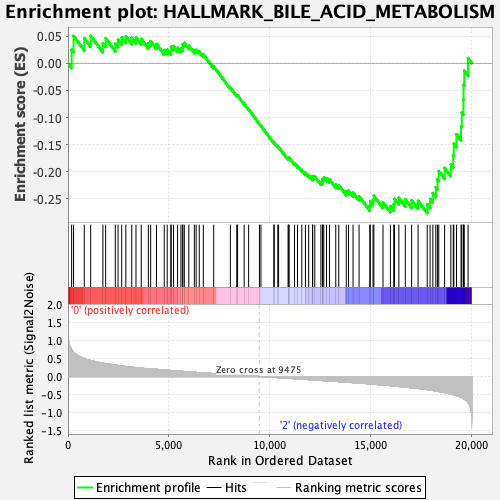
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | GMP.GMP.neu_Pheno.cls #Group1_versus_Group3.GMP.neu_Pheno.cls #Group1_versus_Group3_repos |
| Phenotype | GMP.neu_Pheno.cls#Group1_versus_Group3_repos |
| Upregulated in class | 2 |
| GeneSet | HALLMARK_BILE_ACID_METABOLISM |
| Enrichment Score (ES) | -0.27602693 |
| Normalized Enrichment Score (NES) | -1.1510981 |
| Nominal p-value | 0.2188755 |
| FDR q-value | 0.8697825 |
| FWER p-Value | 0.932 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Pfkm | 180 | 0.750 | 0.0247 | No |
| 2 | Abca8b | 276 | 0.671 | 0.0502 | No |
| 3 | Hao1 | 807 | 0.500 | 0.0461 | No |
| 4 | Abca1 | 1124 | 0.444 | 0.0503 | No |
| 5 | Abca3 | 1731 | 0.371 | 0.0366 | No |
| 6 | Nr1h4 | 1867 | 0.356 | 0.0458 | No |
| 7 | Abca6 | 2346 | 0.319 | 0.0362 | No |
| 8 | Npc1 | 2482 | 0.306 | 0.0432 | No |
| 9 | Abca5 | 2663 | 0.292 | 0.0473 | No |
| 10 | Rbp1 | 2867 | 0.278 | 0.0496 | No |
| 11 | Gnpat | 3162 | 0.260 | 0.0466 | No |
| 12 | Abcd2 | 3371 | 0.246 | 0.0472 | No |
| 13 | Gnmt | 3632 | 0.233 | 0.0447 | No |
| 14 | Slc23a2 | 3986 | 0.217 | 0.0367 | No |
| 15 | Slc35b2 | 4100 | 0.213 | 0.0407 | No |
| 16 | Abca4 | 4387 | 0.199 | 0.0353 | No |
| 17 | Aldh9a1 | 4773 | 0.182 | 0.0242 | No |
| 18 | Slc29a1 | 4920 | 0.175 | 0.0247 | No |
| 19 | Pex19 | 5104 | 0.168 | 0.0231 | No |
| 20 | Akr1d1 | 5111 | 0.167 | 0.0303 | No |
| 21 | Paox | 5232 | 0.161 | 0.0316 | No |
| 22 | Aqp9 | 5434 | 0.153 | 0.0284 | No |
| 23 | Bcar3 | 5600 | 0.146 | 0.0267 | No |
| 24 | Crot | 5687 | 0.143 | 0.0288 | No |
| 25 | Optn | 5690 | 0.142 | 0.0351 | No |
| 26 | Slc22a18 | 5768 | 0.138 | 0.0375 | No |
| 27 | Hsd17b4 | 5992 | 0.129 | 0.0321 | No |
| 28 | Nr3c2 | 6265 | 0.118 | 0.0237 | No |
| 29 | Abcd3 | 6355 | 0.114 | 0.0244 | No |
| 30 | Fdxr | 6508 | 0.108 | 0.0217 | No |
| 31 | Acsl5 | 6712 | 0.100 | 0.0160 | No |
| 32 | Pxmp2 | 7225 | 0.081 | -0.0060 | No |
| 33 | Abca9 | 8052 | 0.052 | -0.0451 | No |
| 34 | Cyp46a1 | 8369 | 0.039 | -0.0592 | No |
| 35 | Dhcr24 | 8410 | 0.038 | -0.0595 | No |
| 36 | Idi1 | 8735 | 0.027 | -0.0746 | No |
| 37 | Gstk1 | 8959 | 0.018 | -0.0849 | No |
| 38 | Gc | 9493 | 0.000 | -0.1117 | No |
| 39 | Lonp2 | 9569 | -0.001 | -0.1154 | No |
| 40 | Abcg4 | 10207 | -0.022 | -0.1464 | No |
| 41 | Abcd1 | 10227 | -0.023 | -0.1463 | No |
| 42 | Hsd17b11 | 10405 | -0.029 | -0.1538 | No |
| 43 | Acsl1 | 10443 | -0.031 | -0.1543 | No |
| 44 | Dio2 | 10914 | -0.048 | -0.1757 | No |
| 45 | Aldh1a1 | 10949 | -0.049 | -0.1752 | No |
| 46 | Fads1 | 10963 | -0.050 | -0.1736 | No |
| 47 | Pex6 | 11234 | -0.059 | -0.1845 | No |
| 48 | Isoc1 | 11380 | -0.063 | -0.1890 | No |
| 49 | Scp2 | 11595 | -0.071 | -0.1965 | No |
| 50 | Pex7 | 11775 | -0.079 | -0.2019 | No |
| 51 | Idh1 | 11935 | -0.085 | -0.2061 | No |
| 52 | Pex11a | 12128 | -0.091 | -0.2116 | No |
| 53 | Cyp7b1 | 12134 | -0.092 | -0.2077 | No |
| 54 | Nedd4 | 12239 | -0.096 | -0.2086 | No |
| 55 | Pex16 | 12542 | -0.108 | -0.2189 | No |
| 56 | Pnpla8 | 12621 | -0.112 | -0.2178 | No |
| 57 | Cyp39a1 | 12628 | -0.112 | -0.2131 | No |
| 58 | Lck | 12684 | -0.114 | -0.2107 | No |
| 59 | Aldh8a1 | 12821 | -0.119 | -0.2122 | No |
| 60 | Slc27a2 | 12967 | -0.122 | -0.2139 | No |
| 61 | Pex1 | 13279 | -0.135 | -0.2235 | No |
| 62 | Atxn1 | 13429 | -0.140 | -0.2246 | No |
| 63 | Ephx2 | 13799 | -0.155 | -0.2362 | No |
| 64 | Ar | 13912 | -0.159 | -0.2346 | No |
| 65 | Fads2 | 14136 | -0.169 | -0.2382 | No |
| 66 | Retsat | 14434 | -0.181 | -0.2450 | No |
| 67 | Cyp27a1 | 14966 | -0.204 | -0.2624 | No |
| 68 | Pex13 | 14989 | -0.205 | -0.2543 | No |
| 69 | Pecr | 15127 | -0.212 | -0.2516 | No |
| 70 | Hsd3b7 | 15169 | -0.214 | -0.2441 | No |
| 71 | Sod1 | 15622 | -0.236 | -0.2561 | No |
| 72 | Slc23a1 | 15989 | -0.253 | -0.2631 | Yes |
| 73 | Gclm | 16155 | -0.262 | -0.2596 | Yes |
| 74 | Pex26 | 16192 | -0.263 | -0.2495 | Yes |
| 75 | Slc27a5 | 16408 | -0.277 | -0.2479 | Yes |
| 76 | Idh2 | 16727 | -0.294 | -0.2506 | Yes |
| 77 | Abca2 | 17042 | -0.313 | -0.2523 | Yes |
| 78 | Cat | 17361 | -0.331 | -0.2533 | Yes |
| 79 | Phyh | 17815 | -0.364 | -0.2597 | Yes |
| 80 | Nudt12 | 17957 | -0.376 | -0.2498 | Yes |
| 81 | Pex11g | 18094 | -0.388 | -0.2392 | Yes |
| 82 | Efhc1 | 18241 | -0.402 | -0.2284 | Yes |
| 83 | Hacl1 | 18321 | -0.410 | -0.2139 | Yes |
| 84 | Rxra | 18388 | -0.416 | -0.1985 | Yes |
| 85 | Bmp6 | 18677 | -0.448 | -0.1928 | Yes |
| 86 | Pex12 | 18986 | -0.486 | -0.1864 | Yes |
| 87 | Lipe | 19106 | -0.504 | -0.1697 | Yes |
| 88 | Soat2 | 19136 | -0.508 | -0.1483 | Yes |
| 89 | Tfcp2l1 | 19266 | -0.532 | -0.1308 | Yes |
| 90 | Mlycd | 19492 | -0.581 | -0.1159 | Yes |
| 91 | Amacr | 19529 | -0.590 | -0.0912 | Yes |
| 92 | Klf1 | 19609 | -0.613 | -0.0676 | Yes |
| 93 | Nr0b2 | 19614 | -0.613 | -0.0401 | Yes |
| 94 | Sult2b1 | 19652 | -0.628 | -0.0137 | Yes |
| 95 | Prdx5 | 19840 | -0.721 | 0.0093 | Yes |