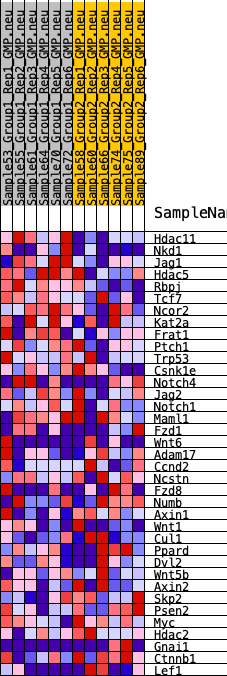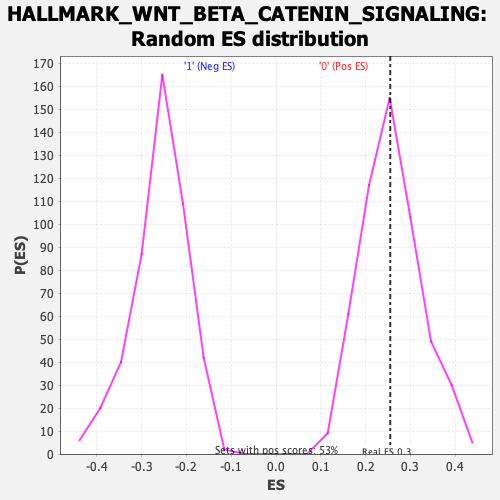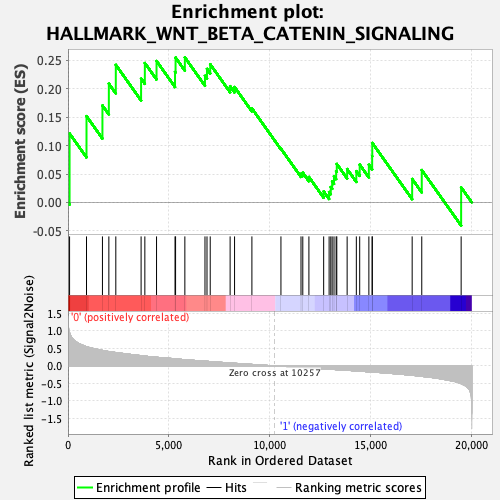
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | GMP.GMP.neu_Pheno.cls #Group1_versus_Group2.GMP.neu_Pheno.cls #Group1_versus_Group2_repos |
| Phenotype | GMP.neu_Pheno.cls#Group1_versus_Group2_repos |
| Upregulated in class | 0 |
| GeneSet | HALLMARK_WNT_BETA_CATENIN_SIGNALING |
| Enrichment Score (ES) | 0.25491905 |
| Normalized Enrichment Score (NES) | 0.9847098 |
| Nominal p-value | 0.51039696 |
| FDR q-value | 0.88224846 |
| FWER p-Value | 0.998 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Hdac11 | 78 | 0.947 | 0.1211 | Yes |
| 2 | Nkd1 | 918 | 0.549 | 0.1517 | Yes |
| 3 | Jag1 | 1708 | 0.441 | 0.1705 | Yes |
| 4 | Hdac5 | 2026 | 0.410 | 0.2087 | Yes |
| 5 | Rbpj | 2371 | 0.381 | 0.2419 | Yes |
| 6 | Tcf7 | 3625 | 0.292 | 0.2177 | Yes |
| 7 | Ncor2 | 3811 | 0.278 | 0.2451 | Yes |
| 8 | Kat2a | 4391 | 0.246 | 0.2486 | Yes |
| 9 | Frat1 | 5308 | 0.202 | 0.2294 | Yes |
| 10 | Ptch1 | 5335 | 0.200 | 0.2546 | Yes |
| 11 | Trp53 | 5795 | 0.176 | 0.2549 | Yes |
| 12 | Csnk1e | 6793 | 0.133 | 0.2227 | No |
| 13 | Notch4 | 6893 | 0.129 | 0.2347 | No |
| 14 | Jag2 | 7053 | 0.122 | 0.2429 | No |
| 15 | Notch1 | 8040 | 0.082 | 0.2043 | No |
| 16 | Maml1 | 8260 | 0.072 | 0.2029 | No |
| 17 | Fzd1 | 9120 | 0.041 | 0.1653 | No |
| 18 | Wnt6 | 10560 | -0.009 | 0.0945 | No |
| 19 | Adam17 | 11555 | -0.044 | 0.0505 | No |
| 20 | Ccnd2 | 11644 | -0.047 | 0.0523 | No |
| 21 | Ncstn | 11947 | -0.058 | 0.0448 | No |
| 22 | Fzd8 | 12679 | -0.085 | 0.0195 | No |
| 23 | Numb | 12949 | -0.094 | 0.0184 | No |
| 24 | Axin1 | 13032 | -0.097 | 0.0272 | No |
| 25 | Wnt1 | 13100 | -0.100 | 0.0370 | No |
| 26 | Cul1 | 13192 | -0.103 | 0.0461 | No |
| 27 | Ppard | 13297 | -0.107 | 0.0550 | No |
| 28 | Dvl2 | 13330 | -0.108 | 0.0677 | No |
| 29 | Wnt5b | 13843 | -0.126 | 0.0588 | No |
| 30 | Axin2 | 14299 | -0.144 | 0.0550 | No |
| 31 | Skp2 | 14466 | -0.150 | 0.0665 | No |
| 32 | Psen2 | 14921 | -0.170 | 0.0663 | No |
| 33 | Myc | 15084 | -0.176 | 0.0815 | No |
| 34 | Hdac2 | 15090 | -0.177 | 0.1046 | No |
| 35 | Gnai1 | 17068 | -0.270 | 0.0413 | No |
| 36 | Ctnnb1 | 17544 | -0.299 | 0.0570 | No |
| 37 | Lef1 | 19497 | -0.508 | 0.0265 | No |