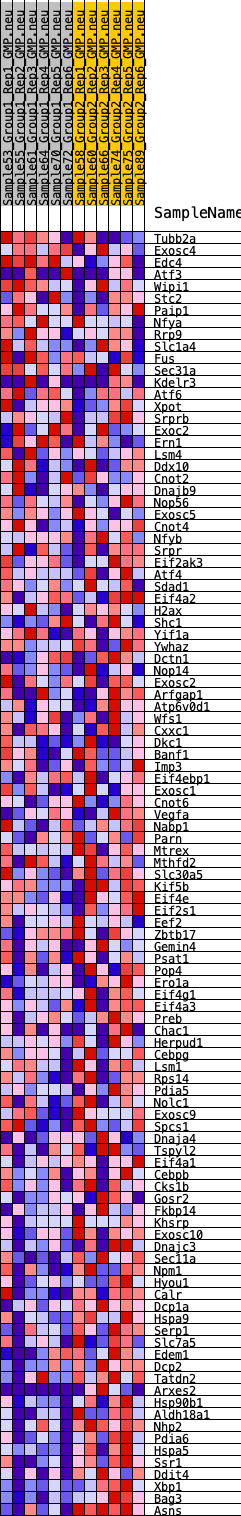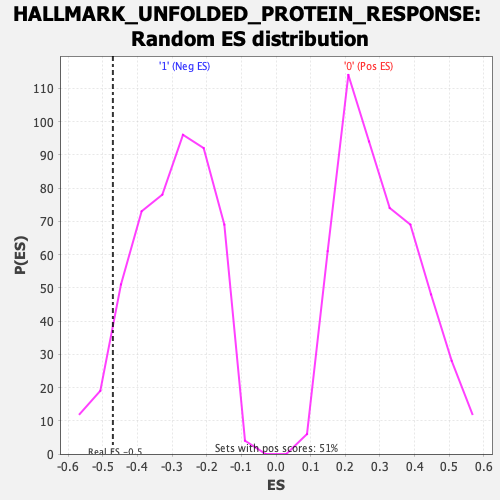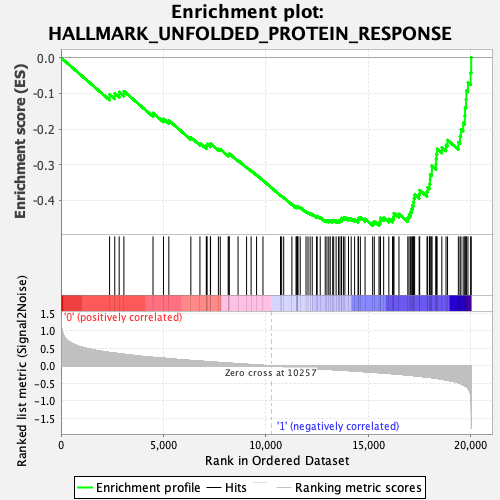
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | GMP.GMP.neu_Pheno.cls #Group1_versus_Group2.GMP.neu_Pheno.cls #Group1_versus_Group2_repos |
| Phenotype | GMP.neu_Pheno.cls#Group1_versus_Group2_repos |
| Upregulated in class | 1 |
| GeneSet | HALLMARK_UNFOLDED_PROTEIN_RESPONSE |
| Enrichment Score (ES) | -0.47053543 |
| Normalized Enrichment Score (NES) | -1.5594841 |
| Nominal p-value | 0.0708502 |
| FDR q-value | 0.12683684 |
| FWER p-Value | 0.25 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Tubb2a | 2370 | 0.381 | -0.1029 | No |
| 2 | Exosc4 | 2623 | 0.363 | -0.1002 | No |
| 3 | Edc4 | 2838 | 0.346 | -0.0963 | No |
| 4 | Atf3 | 3068 | 0.329 | -0.0939 | No |
| 5 | Wipi1 | 4489 | 0.241 | -0.1550 | No |
| 6 | Stc2 | 4999 | 0.217 | -0.1714 | No |
| 7 | Paip1 | 5261 | 0.204 | -0.1759 | No |
| 8 | Nfya | 6335 | 0.152 | -0.2233 | No |
| 9 | Rrp9 | 6778 | 0.134 | -0.2398 | No |
| 10 | Slc1a4 | 7092 | 0.121 | -0.2504 | No |
| 11 | Fus | 7095 | 0.121 | -0.2454 | No |
| 12 | Sec31a | 7128 | 0.119 | -0.2420 | No |
| 13 | Kdelr3 | 7289 | 0.113 | -0.2453 | No |
| 14 | Atf6 | 7296 | 0.113 | -0.2408 | No |
| 15 | Xpot | 7679 | 0.096 | -0.2559 | No |
| 16 | Srprb | 7770 | 0.092 | -0.2565 | No |
| 17 | Exoc2 | 8156 | 0.077 | -0.2726 | No |
| 18 | Ern1 | 8187 | 0.075 | -0.2709 | No |
| 19 | Lsm4 | 8210 | 0.075 | -0.2689 | No |
| 20 | Ddx10 | 8638 | 0.059 | -0.2878 | No |
| 21 | Cnot2 | 9054 | 0.043 | -0.3068 | No |
| 22 | Dnajb9 | 9274 | 0.034 | -0.3164 | No |
| 23 | Nop56 | 9541 | 0.025 | -0.3287 | No |
| 24 | Exosc5 | 9853 | 0.013 | -0.3437 | No |
| 25 | Cnot4 | 10709 | -0.014 | -0.3861 | No |
| 26 | Nfyb | 10765 | -0.017 | -0.3881 | No |
| 27 | Srpr | 10865 | -0.021 | -0.3922 | No |
| 28 | Eif2ak3 | 11265 | -0.034 | -0.4108 | No |
| 29 | Atf4 | 11475 | -0.041 | -0.4196 | No |
| 30 | Sdad1 | 11477 | -0.041 | -0.4179 | No |
| 31 | Eif4a2 | 11539 | -0.043 | -0.4191 | No |
| 32 | H2ax | 11548 | -0.043 | -0.4177 | No |
| 33 | Shc1 | 11558 | -0.044 | -0.4163 | No |
| 34 | Yif1a | 11670 | -0.048 | -0.4198 | No |
| 35 | Ywhaz | 11954 | -0.058 | -0.4316 | No |
| 36 | Dctn1 | 12051 | -0.061 | -0.4338 | No |
| 37 | Nop14 | 12154 | -0.065 | -0.4362 | No |
| 38 | Exosc2 | 12257 | -0.069 | -0.4384 | No |
| 39 | Arfgap1 | 12467 | -0.077 | -0.4457 | No |
| 40 | Atp6v0d1 | 12498 | -0.078 | -0.4439 | No |
| 41 | Wfs1 | 12647 | -0.083 | -0.4478 | No |
| 42 | Cxxc1 | 12890 | -0.092 | -0.4561 | No |
| 43 | Dkc1 | 12975 | -0.095 | -0.4563 | No |
| 44 | Banf1 | 13065 | -0.099 | -0.4566 | No |
| 45 | Imp3 | 13135 | -0.102 | -0.4557 | No |
| 46 | Eif4ebp1 | 13255 | -0.105 | -0.4573 | No |
| 47 | Exosc1 | 13304 | -0.107 | -0.4551 | No |
| 48 | Cnot6 | 13422 | -0.110 | -0.4564 | No |
| 49 | Vegfa | 13536 | -0.114 | -0.4572 | No |
| 50 | Nabp1 | 13608 | -0.118 | -0.4558 | No |
| 51 | Parn | 13682 | -0.120 | -0.4544 | No |
| 52 | Mtrex | 13686 | -0.121 | -0.4494 | No |
| 53 | Mthfd2 | 13778 | -0.124 | -0.4488 | No |
| 54 | Slc30a5 | 13850 | -0.126 | -0.4470 | No |
| 55 | Kif5b | 14026 | -0.133 | -0.4502 | No |
| 56 | Eif4e | 14158 | -0.139 | -0.4509 | No |
| 57 | Eif2s1 | 14324 | -0.145 | -0.4530 | No |
| 58 | Eef2 | 14497 | -0.152 | -0.4553 | No |
| 59 | Zbtb17 | 14506 | -0.152 | -0.4492 | No |
| 60 | Gemin4 | 14605 | -0.156 | -0.4476 | No |
| 61 | Psat1 | 14835 | -0.167 | -0.4520 | No |
| 62 | Pop4 | 15205 | -0.181 | -0.4629 | Yes |
| 63 | Ero1a | 15285 | -0.185 | -0.4590 | Yes |
| 64 | Eif4g1 | 15515 | -0.193 | -0.4624 | Yes |
| 65 | Eif4a3 | 15577 | -0.196 | -0.4572 | Yes |
| 66 | Preb | 15582 | -0.196 | -0.4491 | Yes |
| 67 | Chac1 | 15741 | -0.203 | -0.4484 | Yes |
| 68 | Herpud1 | 15994 | -0.214 | -0.4520 | Yes |
| 69 | Cebpg | 16175 | -0.223 | -0.4517 | Yes |
| 70 | Lsm1 | 16231 | -0.226 | -0.4448 | Yes |
| 71 | Rps14 | 16239 | -0.227 | -0.4356 | Yes |
| 72 | Pdia5 | 16488 | -0.240 | -0.4379 | Yes |
| 73 | Nolc1 | 16920 | -0.262 | -0.4485 | Yes |
| 74 | Exosc9 | 16980 | -0.266 | -0.4402 | Yes |
| 75 | Spcs1 | 17055 | -0.269 | -0.4325 | Yes |
| 76 | Dnaja4 | 17112 | -0.272 | -0.4238 | Yes |
| 77 | Tspyl2 | 17146 | -0.274 | -0.4139 | Yes |
| 78 | Eif4a1 | 17196 | -0.277 | -0.4047 | Yes |
| 79 | Cebpb | 17217 | -0.278 | -0.3940 | Yes |
| 80 | Cks1b | 17253 | -0.281 | -0.3839 | Yes |
| 81 | Gosr2 | 17473 | -0.294 | -0.3824 | Yes |
| 82 | Fkbp14 | 17499 | -0.295 | -0.3712 | Yes |
| 83 | Khsrp | 17857 | -0.321 | -0.3756 | Yes |
| 84 | Exosc10 | 17887 | -0.323 | -0.3634 | Yes |
| 85 | Dnajc3 | 17977 | -0.329 | -0.3540 | Yes |
| 86 | Sec11a | 18003 | -0.331 | -0.3413 | Yes |
| 87 | Npm1 | 18008 | -0.331 | -0.3275 | Yes |
| 88 | Hyou1 | 18092 | -0.337 | -0.3174 | Yes |
| 89 | Calr | 18093 | -0.337 | -0.3032 | Yes |
| 90 | Dcp1a | 18291 | -0.353 | -0.2982 | Yes |
| 91 | Hspa9 | 18298 | -0.354 | -0.2835 | Yes |
| 92 | Serp1 | 18316 | -0.355 | -0.2694 | Yes |
| 93 | Slc7a5 | 18348 | -0.358 | -0.2558 | Yes |
| 94 | Edem1 | 18580 | -0.380 | -0.2513 | Yes |
| 95 | Dcp2 | 18784 | -0.402 | -0.2445 | Yes |
| 96 | Tatdn2 | 18858 | -0.410 | -0.2309 | Yes |
| 97 | Arxes2 | 19385 | -0.483 | -0.2369 | Yes |
| 98 | Hsp90b1 | 19466 | -0.500 | -0.2198 | Yes |
| 99 | Aldh18a1 | 19512 | -0.513 | -0.2004 | Yes |
| 100 | Nhp2 | 19620 | -0.542 | -0.1828 | Yes |
| 101 | Pdia6 | 19702 | -0.567 | -0.1629 | Yes |
| 102 | Hspa5 | 19713 | -0.571 | -0.1393 | Yes |
| 103 | Ssr1 | 19759 | -0.584 | -0.1169 | Yes |
| 104 | Ddit4 | 19775 | -0.595 | -0.0925 | Yes |
| 105 | Xbp1 | 19854 | -0.641 | -0.0694 | Yes |
| 106 | Bag3 | 19985 | -0.807 | -0.0418 | Yes |
| 107 | Asns | 20013 | -1.036 | 0.0007 | Yes |