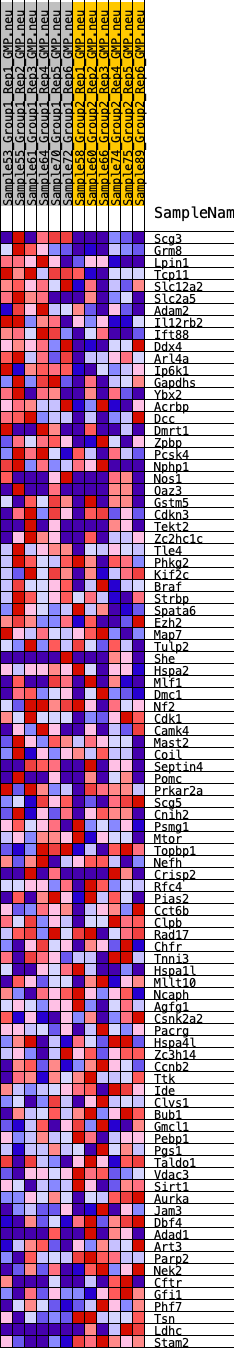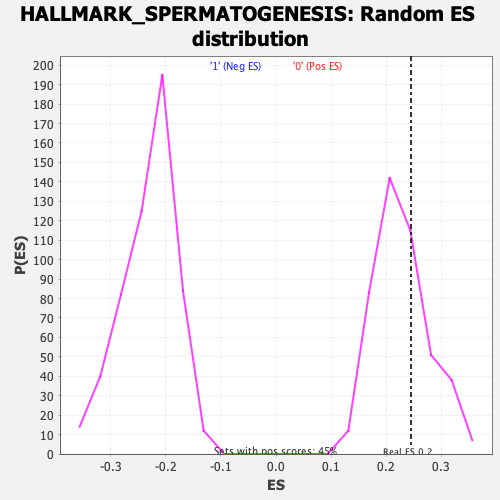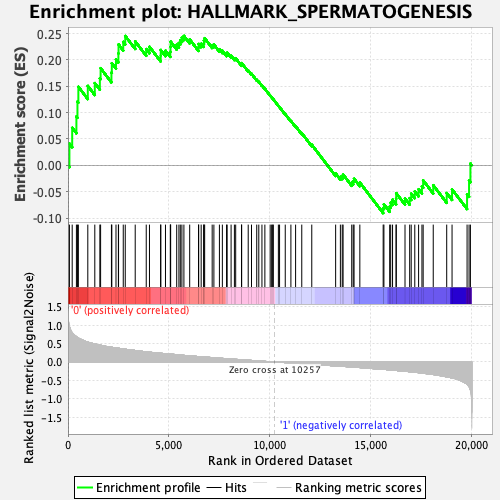
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | GMP.GMP.neu_Pheno.cls #Group1_versus_Group2.GMP.neu_Pheno.cls #Group1_versus_Group2_repos |
| Phenotype | GMP.neu_Pheno.cls#Group1_versus_Group2_repos |
| Upregulated in class | 0 |
| GeneSet | HALLMARK_SPERMATOGENESIS |
| Enrichment Score (ES) | 0.24548872 |
| Normalized Enrichment Score (NES) | 1.0745268 |
| Nominal p-value | 0.3549107 |
| FDR q-value | 1.0 |
| FWER p-Value | 0.985 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Scg3 | 70 | 0.960 | 0.0412 | Yes |
| 2 | Grm8 | 208 | 0.792 | 0.0712 | Yes |
| 3 | Lpin1 | 417 | 0.685 | 0.0927 | Yes |
| 4 | Tcp11 | 469 | 0.662 | 0.1210 | Yes |
| 5 | Slc12a2 | 514 | 0.648 | 0.1490 | Yes |
| 6 | Slc2a5 | 982 | 0.537 | 0.1506 | Yes |
| 7 | Adam2 | 1329 | 0.488 | 0.1560 | Yes |
| 8 | Il12rb2 | 1580 | 0.459 | 0.1648 | Yes |
| 9 | Ift88 | 1617 | 0.454 | 0.1842 | Yes |
| 10 | Ddx4 | 2154 | 0.400 | 0.1759 | Yes |
| 11 | Arl4a | 2173 | 0.398 | 0.1935 | Yes |
| 12 | Ip6k1 | 2380 | 0.381 | 0.2009 | Yes |
| 13 | Gapdhs | 2499 | 0.371 | 0.2123 | Yes |
| 14 | Ybx2 | 2507 | 0.371 | 0.2293 | Yes |
| 15 | Acrbp | 2740 | 0.354 | 0.2341 | Yes |
| 16 | Dcc | 2839 | 0.346 | 0.2453 | Yes |
| 17 | Dmrt1 | 3333 | 0.311 | 0.2351 | Yes |
| 18 | Zpbp | 3880 | 0.274 | 0.2205 | Yes |
| 19 | Pcsk4 | 4040 | 0.265 | 0.2248 | Yes |
| 20 | Nphp1 | 4597 | 0.235 | 0.2079 | Yes |
| 21 | Nos1 | 4599 | 0.235 | 0.2188 | Yes |
| 22 | Oaz3 | 4834 | 0.224 | 0.2174 | Yes |
| 23 | Gstm5 | 5073 | 0.214 | 0.2155 | Yes |
| 24 | Cdkn3 | 5083 | 0.213 | 0.2249 | Yes |
| 25 | Tekt2 | 5091 | 0.213 | 0.2345 | Yes |
| 26 | Zc2hc1c | 5391 | 0.197 | 0.2287 | Yes |
| 27 | Tle4 | 5501 | 0.191 | 0.2322 | Yes |
| 28 | Phkg2 | 5569 | 0.187 | 0.2375 | Yes |
| 29 | Kif2c | 5648 | 0.183 | 0.2421 | Yes |
| 30 | Braf | 5748 | 0.179 | 0.2455 | Yes |
| 31 | Strbp | 6035 | 0.164 | 0.2388 | No |
| 32 | Spata6 | 6473 | 0.146 | 0.2237 | No |
| 33 | Ezh2 | 6481 | 0.146 | 0.2301 | No |
| 34 | Map7 | 6604 | 0.141 | 0.2306 | No |
| 35 | Tulp2 | 6727 | 0.137 | 0.2308 | No |
| 36 | She | 6742 | 0.136 | 0.2365 | No |
| 37 | Hspa2 | 6777 | 0.134 | 0.2410 | No |
| 38 | Mlf1 | 7150 | 0.118 | 0.2279 | No |
| 39 | Dmc1 | 7235 | 0.115 | 0.2290 | No |
| 40 | Nf2 | 7509 | 0.103 | 0.2201 | No |
| 41 | Cdk1 | 7655 | 0.097 | 0.2174 | No |
| 42 | Camk4 | 7863 | 0.089 | 0.2111 | No |
| 43 | Mast2 | 7892 | 0.088 | 0.2138 | No |
| 44 | Coil | 8088 | 0.080 | 0.2077 | No |
| 45 | Septin4 | 8252 | 0.072 | 0.2029 | No |
| 46 | Pomc | 8316 | 0.070 | 0.2031 | No |
| 47 | Prkar2a | 8604 | 0.061 | 0.1915 | No |
| 48 | Scg5 | 8613 | 0.060 | 0.1939 | No |
| 49 | Cnih2 | 8937 | 0.048 | 0.1799 | No |
| 50 | Psmg1 | 9100 | 0.041 | 0.1737 | No |
| 51 | Mtor | 9360 | 0.031 | 0.1622 | No |
| 52 | Topbp1 | 9462 | 0.027 | 0.1584 | No |
| 53 | Nefh | 9618 | 0.022 | 0.1516 | No |
| 54 | Crisp2 | 9763 | 0.017 | 0.1452 | No |
| 55 | Rfc4 | 10026 | 0.008 | 0.1324 | No |
| 56 | Pias2 | 10100 | 0.006 | 0.1290 | No |
| 57 | Cct6b | 10148 | 0.004 | 0.1268 | No |
| 58 | Clpb | 10183 | 0.002 | 0.1252 | No |
| 59 | Rad17 | 10433 | -0.005 | 0.1129 | No |
| 60 | Chfr | 10492 | -0.006 | 0.1103 | No |
| 61 | Tnni3 | 10774 | -0.017 | 0.0970 | No |
| 62 | Hspa1l | 11054 | -0.026 | 0.0842 | No |
| 63 | Mllt10 | 11287 | -0.035 | 0.0742 | No |
| 64 | Ncaph | 11593 | -0.045 | 0.0610 | No |
| 65 | Agfg1 | 12089 | -0.062 | 0.0391 | No |
| 66 | Csnk2a2 | 13273 | -0.106 | -0.0153 | No |
| 67 | Pacrg | 13508 | -0.114 | -0.0218 | No |
| 68 | Hspa4l | 13599 | -0.117 | -0.0208 | No |
| 69 | Zc3h14 | 13642 | -0.119 | -0.0174 | No |
| 70 | Ccnb2 | 14071 | -0.135 | -0.0326 | No |
| 71 | Ttk | 14144 | -0.138 | -0.0297 | No |
| 72 | Ide | 14186 | -0.140 | -0.0253 | No |
| 73 | Clvs1 | 14473 | -0.150 | -0.0326 | No |
| 74 | Bub1 | 15630 | -0.198 | -0.0814 | No |
| 75 | Gmcl1 | 15667 | -0.200 | -0.0738 | No |
| 76 | Pebp1 | 15953 | -0.212 | -0.0783 | No |
| 77 | Pgs1 | 16003 | -0.215 | -0.0707 | No |
| 78 | Taldo1 | 16095 | -0.220 | -0.0650 | No |
| 79 | Vdac3 | 16272 | -0.229 | -0.0632 | No |
| 80 | Sirt1 | 16277 | -0.229 | -0.0528 | No |
| 81 | Aurka | 16713 | -0.251 | -0.0629 | No |
| 82 | Jam3 | 16945 | -0.264 | -0.0622 | No |
| 83 | Dbf4 | 17020 | -0.268 | -0.0534 | No |
| 84 | Adad1 | 17198 | -0.277 | -0.0494 | No |
| 85 | Art3 | 17387 | -0.289 | -0.0454 | No |
| 86 | Parp2 | 17553 | -0.300 | -0.0397 | No |
| 87 | Nek2 | 17613 | -0.305 | -0.0284 | No |
| 88 | Cftr | 18113 | -0.338 | -0.0377 | No |
| 89 | Gfi1 | 18781 | -0.402 | -0.0524 | No |
| 90 | Phf7 | 19046 | -0.431 | -0.0456 | No |
| 91 | Tsn | 19791 | -0.601 | -0.0549 | No |
| 92 | Ldhc | 19890 | -0.667 | -0.0287 | No |
| 93 | Stam2 | 19957 | -0.762 | 0.0035 | No |