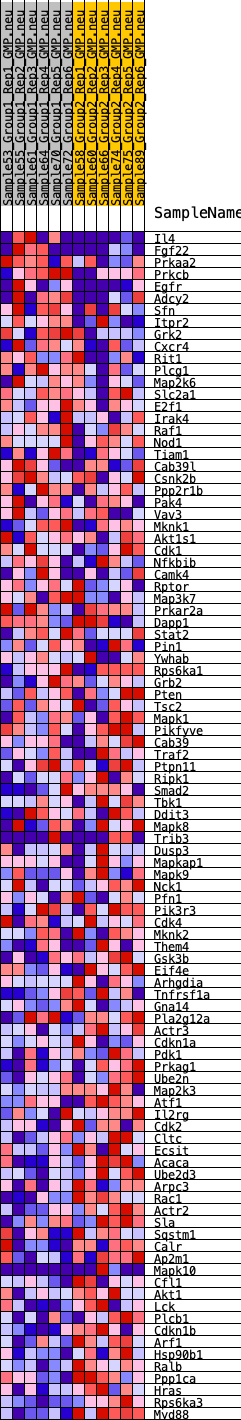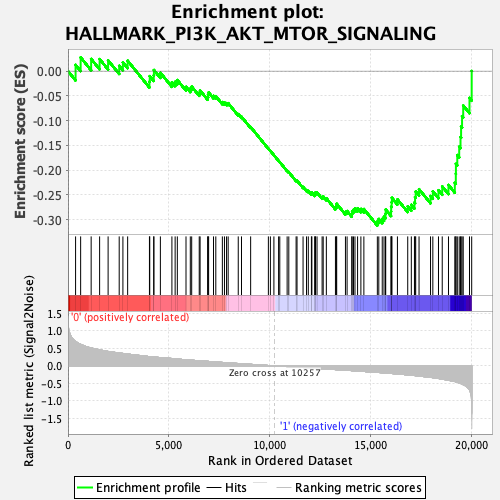
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | GMP.GMP.neu_Pheno.cls #Group1_versus_Group2.GMP.neu_Pheno.cls #Group1_versus_Group2_repos |
| Phenotype | GMP.neu_Pheno.cls#Group1_versus_Group2_repos |
| Upregulated in class | 1 |
| GeneSet | HALLMARK_PI3K_AKT_MTOR_SIGNALING |
| Enrichment Score (ES) | -0.31230143 |
| Normalized Enrichment Score (NES) | -1.6306797 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.1345492 |
| FWER p-Value | 0.178 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Il4 | 376 | 0.700 | 0.0126 | No |
| 2 | Fgf22 | 627 | 0.616 | 0.0277 | No |
| 3 | Prkaa2 | 1148 | 0.512 | 0.0246 | No |
| 4 | Prkcb | 1568 | 0.461 | 0.0243 | No |
| 5 | Egfr | 1989 | 0.412 | 0.0218 | No |
| 6 | Adcy2 | 2539 | 0.369 | 0.0108 | No |
| 7 | Sfn | 2723 | 0.356 | 0.0176 | No |
| 8 | Itpr2 | 2958 | 0.337 | 0.0210 | No |
| 9 | Grk2 | 4039 | 0.265 | -0.0213 | No |
| 10 | Cxcr4 | 4054 | 0.265 | -0.0101 | No |
| 11 | Rit1 | 4251 | 0.252 | -0.0086 | No |
| 12 | Plcg1 | 4260 | 0.252 | 0.0023 | No |
| 13 | Map2k6 | 4581 | 0.235 | -0.0032 | No |
| 14 | Slc2a1 | 5153 | 0.209 | -0.0225 | No |
| 15 | E2f1 | 5313 | 0.202 | -0.0214 | No |
| 16 | Irak4 | 5422 | 0.196 | -0.0180 | No |
| 17 | Raf1 | 5852 | 0.173 | -0.0318 | No |
| 18 | Nod1 | 6065 | 0.163 | -0.0351 | No |
| 19 | Tiam1 | 6134 | 0.161 | -0.0313 | No |
| 20 | Cab39l | 6508 | 0.145 | -0.0435 | No |
| 21 | Csnk2b | 6551 | 0.144 | -0.0391 | No |
| 22 | Ppp2r1b | 6925 | 0.128 | -0.0521 | No |
| 23 | Pak4 | 6948 | 0.127 | -0.0475 | No |
| 24 | Vav3 | 6974 | 0.126 | -0.0431 | No |
| 25 | Mknk1 | 7216 | 0.116 | -0.0500 | No |
| 26 | Akt1s1 | 7330 | 0.111 | -0.0507 | No |
| 27 | Cdk1 | 7655 | 0.097 | -0.0626 | No |
| 28 | Nfkbib | 7759 | 0.093 | -0.0636 | No |
| 29 | Camk4 | 7863 | 0.089 | -0.0647 | No |
| 30 | Rptor | 7943 | 0.086 | -0.0649 | No |
| 31 | Map3k7 | 8443 | 0.066 | -0.0869 | No |
| 32 | Prkar2a | 8604 | 0.061 | -0.0922 | No |
| 33 | Dapp1 | 9058 | 0.043 | -0.1130 | No |
| 34 | Stat2 | 9937 | 0.010 | -0.1566 | No |
| 35 | Pin1 | 10037 | 0.007 | -0.1613 | No |
| 36 | Ywhab | 10211 | 0.001 | -0.1699 | No |
| 37 | Rps6ka1 | 10444 | -0.005 | -0.1813 | No |
| 38 | Grb2 | 10503 | -0.007 | -0.1839 | No |
| 39 | Pten | 10867 | -0.021 | -0.2012 | No |
| 40 | Tsc2 | 10950 | -0.023 | -0.2043 | No |
| 41 | Mapk1 | 11312 | -0.036 | -0.2208 | No |
| 42 | Pikfyve | 11358 | -0.038 | -0.2214 | No |
| 43 | Cab39 | 11657 | -0.047 | -0.2342 | No |
| 44 | Traf2 | 11829 | -0.054 | -0.2403 | No |
| 45 | Ptpn11 | 11917 | -0.057 | -0.2422 | No |
| 46 | Ripk1 | 12071 | -0.062 | -0.2471 | No |
| 47 | Smad2 | 12094 | -0.063 | -0.2453 | No |
| 48 | Tbk1 | 12238 | -0.068 | -0.2495 | No |
| 49 | Ddit3 | 12250 | -0.068 | -0.2469 | No |
| 50 | Mapk8 | 12275 | -0.069 | -0.2450 | No |
| 51 | Trib3 | 12350 | -0.072 | -0.2455 | No |
| 52 | Dusp3 | 12602 | -0.082 | -0.2544 | No |
| 53 | Mapkap1 | 12656 | -0.084 | -0.2533 | No |
| 54 | Mapk9 | 12812 | -0.090 | -0.2571 | No |
| 55 | Nck1 | 13263 | -0.106 | -0.2749 | No |
| 56 | Pfn1 | 13307 | -0.107 | -0.2722 | No |
| 57 | Pik3r3 | 13325 | -0.108 | -0.2682 | No |
| 58 | Cdk4 | 13759 | -0.123 | -0.2845 | No |
| 59 | Mknk2 | 13841 | -0.126 | -0.2829 | No |
| 60 | Them4 | 14073 | -0.135 | -0.2884 | No |
| 61 | Gsk3b | 14091 | -0.136 | -0.2831 | No |
| 62 | Eif4e | 14158 | -0.139 | -0.2802 | No |
| 63 | Arhgdia | 14232 | -0.142 | -0.2775 | No |
| 64 | Tnfrsf1a | 14361 | -0.147 | -0.2773 | No |
| 65 | Gna14 | 14525 | -0.153 | -0.2786 | No |
| 66 | Pla2g12a | 14673 | -0.160 | -0.2788 | No |
| 67 | Actr3 | 15341 | -0.188 | -0.3039 | Yes |
| 68 | Cdkn1a | 15413 | -0.189 | -0.2989 | Yes |
| 69 | Pdk1 | 15584 | -0.196 | -0.2986 | Yes |
| 70 | Prkag1 | 15672 | -0.200 | -0.2940 | Yes |
| 71 | Ube2n | 15747 | -0.203 | -0.2886 | Yes |
| 72 | Map2k3 | 15755 | -0.203 | -0.2798 | Yes |
| 73 | Atf1 | 16021 | -0.216 | -0.2834 | Yes |
| 74 | Il2rg | 16027 | -0.216 | -0.2740 | Yes |
| 75 | Cdk2 | 16050 | -0.217 | -0.2653 | Yes |
| 76 | Cltc | 16062 | -0.218 | -0.2561 | Yes |
| 77 | Ecsit | 16337 | -0.232 | -0.2594 | Yes |
| 78 | Acaca | 16848 | -0.258 | -0.2734 | Yes |
| 79 | Ube2d3 | 17026 | -0.268 | -0.2702 | Yes |
| 80 | Arpc3 | 17181 | -0.276 | -0.2656 | Yes |
| 81 | Rac1 | 17212 | -0.278 | -0.2546 | Yes |
| 82 | Actr2 | 17243 | -0.280 | -0.2435 | Yes |
| 83 | Sla | 17409 | -0.290 | -0.2388 | Yes |
| 84 | Sqstm1 | 17979 | -0.329 | -0.2525 | Yes |
| 85 | Calr | 18093 | -0.337 | -0.2430 | Yes |
| 86 | Ap2m1 | 18373 | -0.361 | -0.2408 | Yes |
| 87 | Mapk10 | 18559 | -0.379 | -0.2331 | Yes |
| 88 | Cfl1 | 18869 | -0.411 | -0.2301 | Yes |
| 89 | Akt1 | 19183 | -0.448 | -0.2257 | Yes |
| 90 | Lck | 19233 | -0.456 | -0.2077 | Yes |
| 91 | Plcb1 | 19238 | -0.457 | -0.1873 | Yes |
| 92 | Cdkn1b | 19306 | -0.468 | -0.1697 | Yes |
| 93 | Arf1 | 19401 | -0.485 | -0.1526 | Yes |
| 94 | Hsp90b1 | 19466 | -0.500 | -0.1334 | Yes |
| 95 | Ralb | 19495 | -0.508 | -0.1119 | Yes |
| 96 | Ppp1ca | 19543 | -0.521 | -0.0909 | Yes |
| 97 | Hras | 19600 | -0.538 | -0.0695 | Yes |
| 98 | Rps6ka3 | 19911 | -0.688 | -0.0542 | Yes |
| 99 | Myd88 | 20020 | -1.333 | 0.0003 | Yes |