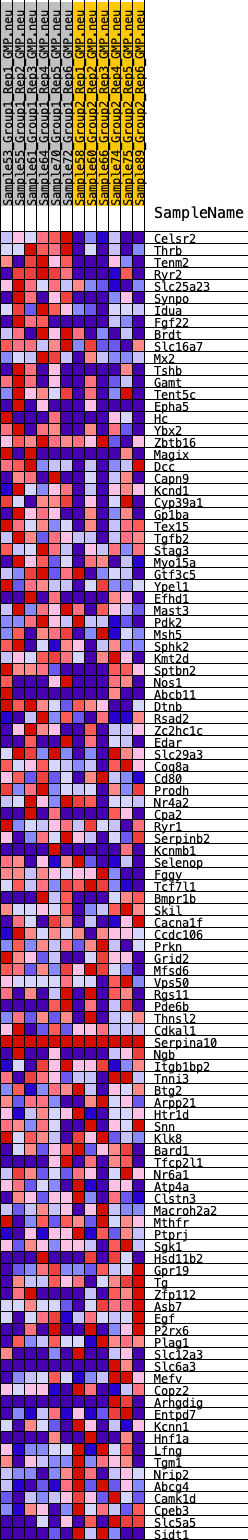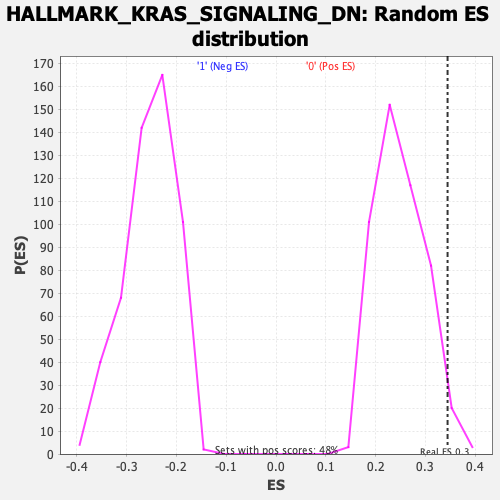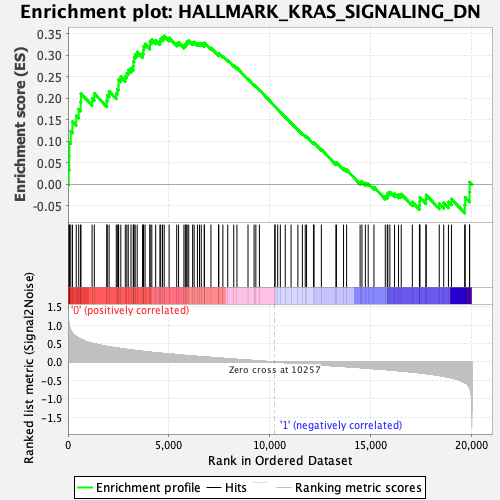
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | GMP.GMP.neu_Pheno.cls #Group1_versus_Group2.GMP.neu_Pheno.cls #Group1_versus_Group2_repos |
| Phenotype | GMP.neu_Pheno.cls#Group1_versus_Group2_repos |
| Upregulated in class | 0 |
| GeneSet | HALLMARK_KRAS_SIGNALING_DN |
| Enrichment Score (ES) | 0.34483966 |
| Normalized Enrichment Score (NES) | 1.3812003 |
| Nominal p-value | 0.027196653 |
| FDR q-value | 0.8383489 |
| FWER p-Value | 0.599 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Celsr2 | 36 | 1.061 | 0.0339 | Yes |
| 2 | Thrb | 62 | 0.982 | 0.0656 | Yes |
| 3 | Tenm2 | 66 | 0.975 | 0.0983 | Yes |
| 4 | Ryr2 | 137 | 0.857 | 0.1235 | Yes |
| 5 | Slc25a23 | 220 | 0.785 | 0.1458 | Yes |
| 6 | Synpo | 408 | 0.688 | 0.1596 | Yes |
| 7 | Idua | 532 | 0.642 | 0.1750 | Yes |
| 8 | Fgf22 | 627 | 0.616 | 0.1910 | Yes |
| 9 | Brdt | 639 | 0.613 | 0.2110 | Yes |
| 10 | Slc16a7 | 1195 | 0.505 | 0.2002 | Yes |
| 11 | Mx2 | 1305 | 0.492 | 0.2112 | Yes |
| 12 | Tshb | 1921 | 0.418 | 0.1944 | Yes |
| 13 | Gamt | 1945 | 0.417 | 0.2073 | Yes |
| 14 | Tent5c | 2038 | 0.409 | 0.2164 | Yes |
| 15 | Epha5 | 2394 | 0.379 | 0.2113 | Yes |
| 16 | Hc | 2445 | 0.376 | 0.2214 | Yes |
| 17 | Ybx2 | 2507 | 0.371 | 0.2308 | Yes |
| 18 | Zbtb16 | 2508 | 0.371 | 0.2433 | Yes |
| 19 | Magix | 2620 | 0.363 | 0.2500 | Yes |
| 20 | Dcc | 2839 | 0.346 | 0.2506 | Yes |
| 21 | Capn9 | 2917 | 0.340 | 0.2582 | Yes |
| 22 | Kcnd1 | 2988 | 0.335 | 0.2659 | Yes |
| 23 | Cyp39a1 | 3125 | 0.325 | 0.2700 | Yes |
| 24 | Gp1ba | 3239 | 0.318 | 0.2751 | Yes |
| 25 | Tex15 | 3245 | 0.317 | 0.2855 | Yes |
| 26 | Tgfb2 | 3275 | 0.315 | 0.2946 | Yes |
| 27 | Stag3 | 3331 | 0.311 | 0.3023 | Yes |
| 28 | Myo15a | 3431 | 0.305 | 0.3076 | Yes |
| 29 | Gtf3c5 | 3697 | 0.286 | 0.3039 | Yes |
| 30 | Ypel1 | 3734 | 0.283 | 0.3116 | Yes |
| 31 | Efhd1 | 3748 | 0.281 | 0.3204 | Yes |
| 32 | Mast3 | 3822 | 0.277 | 0.3260 | Yes |
| 33 | Pdk2 | 4064 | 0.264 | 0.3228 | Yes |
| 34 | Msh5 | 4069 | 0.264 | 0.3315 | Yes |
| 35 | Sphk2 | 4156 | 0.259 | 0.3359 | Yes |
| 36 | Kmt2d | 4348 | 0.247 | 0.3346 | Yes |
| 37 | Sptbn2 | 4558 | 0.236 | 0.3321 | Yes |
| 38 | Nos1 | 4599 | 0.235 | 0.3380 | Yes |
| 39 | Abcb11 | 4688 | 0.231 | 0.3413 | Yes |
| 40 | Dtnb | 4771 | 0.227 | 0.3448 | Yes |
| 41 | Rsad2 | 5014 | 0.216 | 0.3400 | No |
| 42 | Zc2hc1c | 5391 | 0.197 | 0.3277 | No |
| 43 | Edar | 5483 | 0.192 | 0.3296 | No |
| 44 | Slc29a3 | 5749 | 0.179 | 0.3223 | No |
| 45 | Coq8a | 5817 | 0.175 | 0.3248 | No |
| 46 | Cd80 | 5865 | 0.172 | 0.3283 | No |
| 47 | Prodh | 5909 | 0.171 | 0.3319 | No |
| 48 | Nr4a2 | 5987 | 0.166 | 0.3336 | No |
| 49 | Cpa2 | 6181 | 0.159 | 0.3292 | No |
| 50 | Ryr1 | 6255 | 0.156 | 0.3308 | No |
| 51 | Serpinb2 | 6419 | 0.148 | 0.3276 | No |
| 52 | Kcnmb1 | 6521 | 0.144 | 0.3274 | No |
| 53 | Selenop | 6609 | 0.141 | 0.3278 | No |
| 54 | Fggy | 6755 | 0.135 | 0.3250 | No |
| 55 | Tcf7l1 | 6769 | 0.135 | 0.3289 | No |
| 56 | Bmpr1b | 7090 | 0.121 | 0.3169 | No |
| 57 | Skil | 7465 | 0.105 | 0.3016 | No |
| 58 | Cacna1f | 7479 | 0.104 | 0.3045 | No |
| 59 | Ccdc106 | 7680 | 0.096 | 0.2977 | No |
| 60 | Prkn | 7921 | 0.086 | 0.2885 | No |
| 61 | Grid2 | 8219 | 0.074 | 0.2761 | No |
| 62 | Mfsd6 | 8379 | 0.069 | 0.2704 | No |
| 63 | Vps50 | 8927 | 0.048 | 0.2446 | No |
| 64 | Rgs11 | 9228 | 0.036 | 0.2308 | No |
| 65 | Pde6b | 9316 | 0.033 | 0.2275 | No |
| 66 | Thnsl2 | 9492 | 0.026 | 0.2196 | No |
| 67 | Cdkal1 | 10256 | 0.000 | 0.1813 | No |
| 68 | Serpina10 | 10268 | 0.000 | 0.1807 | No |
| 69 | Ngb | 10399 | -0.004 | 0.1743 | No |
| 70 | Itgb1bp2 | 10530 | -0.008 | 0.1681 | No |
| 71 | Tnni3 | 10774 | -0.017 | 0.1564 | No |
| 72 | Btg2 | 11063 | -0.026 | 0.1429 | No |
| 73 | Arpp21 | 11400 | -0.039 | 0.1273 | No |
| 74 | Htr1d | 11623 | -0.046 | 0.1177 | No |
| 75 | Snn | 11765 | -0.052 | 0.1124 | No |
| 76 | Klk8 | 11831 | -0.054 | 0.1109 | No |
| 77 | Bard1 | 12174 | -0.066 | 0.0960 | No |
| 78 | Tfcp2l1 | 12202 | -0.067 | 0.0969 | No |
| 79 | Nr6a1 | 12565 | -0.080 | 0.0814 | No |
| 80 | Atp4a | 13286 | -0.107 | 0.0488 | No |
| 81 | Clstn3 | 13313 | -0.107 | 0.0511 | No |
| 82 | Macroh2a2 | 13665 | -0.120 | 0.0375 | No |
| 83 | Mthfr | 13817 | -0.125 | 0.0342 | No |
| 84 | Ptprj | 14486 | -0.151 | 0.0057 | No |
| 85 | Sgk1 | 14577 | -0.156 | 0.0064 | No |
| 86 | Hsd11b2 | 14748 | -0.163 | 0.0034 | No |
| 87 | Gpr19 | 14888 | -0.169 | 0.0021 | No |
| 88 | Tg | 15173 | -0.180 | -0.0061 | No |
| 89 | Zfp112 | 15735 | -0.202 | -0.0275 | No |
| 90 | Asb7 | 15833 | -0.207 | -0.0254 | No |
| 91 | Egf | 15861 | -0.208 | -0.0198 | No |
| 92 | P2rx6 | 15962 | -0.213 | -0.0177 | No |
| 93 | Plag1 | 16193 | -0.225 | -0.0217 | No |
| 94 | Slc12a3 | 16392 | -0.235 | -0.0237 | No |
| 95 | Slc6a3 | 16526 | -0.241 | -0.0222 | No |
| 96 | Mefv | 17077 | -0.270 | -0.0408 | No |
| 97 | Copz2 | 17428 | -0.291 | -0.0486 | No |
| 98 | Arhgdig | 17441 | -0.292 | -0.0393 | No |
| 99 | Entpd7 | 17450 | -0.292 | -0.0299 | No |
| 100 | Kcnn1 | 17750 | -0.313 | -0.0344 | No |
| 101 | Hnf1a | 17763 | -0.314 | -0.0244 | No |
| 102 | Lfng | 18412 | -0.364 | -0.0447 | No |
| 103 | Tgm1 | 18632 | -0.386 | -0.0427 | No |
| 104 | Nrip2 | 18867 | -0.411 | -0.0406 | No |
| 105 | Abcg4 | 19016 | -0.428 | -0.0337 | No |
| 106 | Camk1d | 19674 | -0.557 | -0.0479 | No |
| 107 | Cpeb3 | 19698 | -0.567 | -0.0300 | No |
| 108 | Slc5a5 | 19913 | -0.689 | -0.0176 | No |
| 109 | Sidt1 | 19918 | -0.691 | 0.0054 | No |