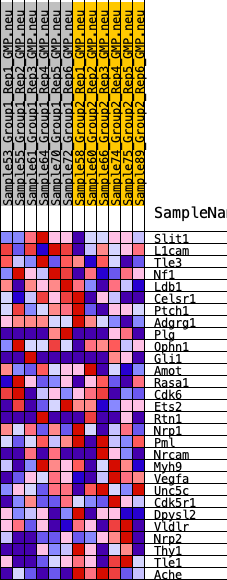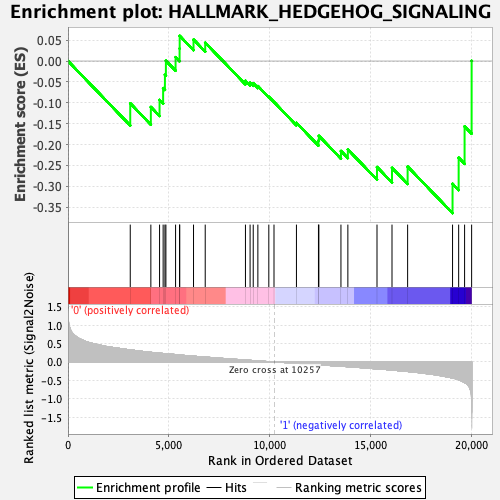
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | GMP.GMP.neu_Pheno.cls #Group1_versus_Group2.GMP.neu_Pheno.cls #Group1_versus_Group2_repos |
| Phenotype | GMP.neu_Pheno.cls#Group1_versus_Group2_repos |
| Upregulated in class | 1 |
| GeneSet | HALLMARK_HEDGEHOG_SIGNALING |
| Enrichment Score (ES) | -0.3641253 |
| Normalized Enrichment Score (NES) | -1.1521693 |
| Nominal p-value | 0.2661448 |
| FDR q-value | 0.5589607 |
| FWER p-Value | 0.947 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Slit1 | 3087 | 0.329 | -0.1013 | No |
| 2 | L1cam | 4109 | 0.262 | -0.1100 | No |
| 3 | Tle3 | 4541 | 0.238 | -0.0932 | No |
| 4 | Nf1 | 4718 | 0.230 | -0.0649 | No |
| 5 | Ldb1 | 4807 | 0.225 | -0.0329 | No |
| 6 | Celsr1 | 4855 | 0.223 | 0.0007 | No |
| 7 | Ptch1 | 5335 | 0.200 | 0.0092 | No |
| 8 | Adgrg1 | 5534 | 0.190 | 0.0299 | No |
| 9 | Plg | 5539 | 0.189 | 0.0602 | No |
| 10 | Ophn1 | 6225 | 0.157 | 0.0513 | No |
| 11 | Gli1 | 6805 | 0.133 | 0.0439 | No |
| 12 | Amot | 8800 | 0.053 | -0.0473 | No |
| 13 | Rasa1 | 9030 | 0.044 | -0.0516 | No |
| 14 | Cdk6 | 9186 | 0.038 | -0.0533 | No |
| 15 | Ets2 | 9416 | 0.029 | -0.0600 | No |
| 16 | Rtn1 | 9959 | 0.009 | -0.0856 | No |
| 17 | Nrp1 | 10216 | 0.001 | -0.0983 | No |
| 18 | Pml | 11329 | -0.037 | -0.1479 | No |
| 19 | Nrcam | 12421 | -0.075 | -0.1904 | No |
| 20 | Myh9 | 12441 | -0.076 | -0.1791 | No |
| 21 | Vegfa | 13536 | -0.114 | -0.2153 | No |
| 22 | Unc5c | 13879 | -0.128 | -0.2118 | No |
| 23 | Cdk5r1 | 15324 | -0.187 | -0.2538 | No |
| 24 | Dpysl2 | 16068 | -0.218 | -0.2557 | No |
| 25 | Vldlr | 16843 | -0.258 | -0.2528 | No |
| 26 | Nrp2 | 19071 | -0.435 | -0.2939 | Yes |
| 27 | Thy1 | 19374 | -0.480 | -0.2315 | Yes |
| 28 | Tle1 | 19668 | -0.555 | -0.1564 | Yes |
| 29 | Ache | 20017 | -1.079 | 0.0004 | Yes |