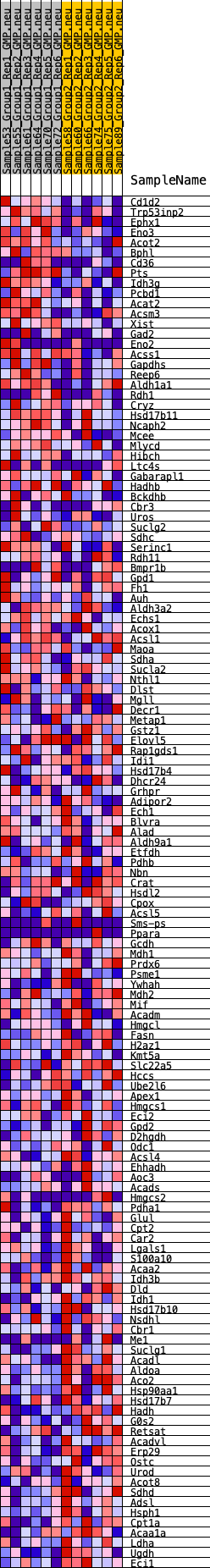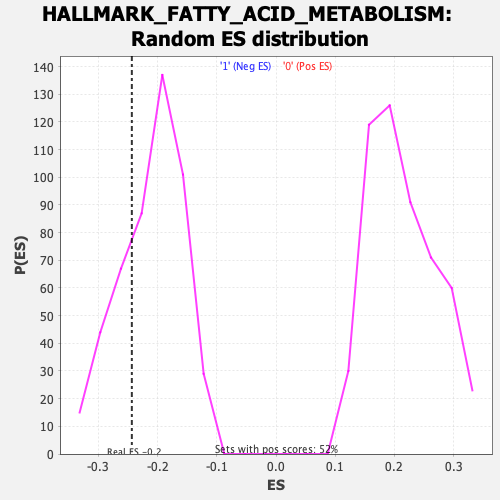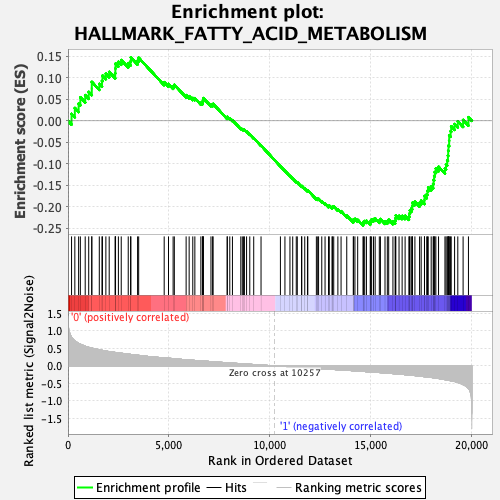
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | GMP.GMP.neu_Pheno.cls #Group1_versus_Group2.GMP.neu_Pheno.cls #Group1_versus_Group2_repos |
| Phenotype | GMP.neu_Pheno.cls#Group1_versus_Group2_repos |
| Upregulated in class | 1 |
| GeneSet | HALLMARK_FATTY_ACID_METABOLISM |
| Enrichment Score (ES) | -0.24333687 |
| Normalized Enrichment Score (NES) | -1.1539226 |
| Nominal p-value | 0.27916667 |
| FDR q-value | 0.5975884 |
| FWER p-Value | 0.947 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Cd1d2 | 176 | 0.818 | 0.0161 | No |
| 2 | Trp53inp2 | 339 | 0.717 | 0.0298 | No |
| 3 | Ephx1 | 527 | 0.643 | 0.0400 | No |
| 4 | Eno3 | 612 | 0.621 | 0.0547 | No |
| 5 | Acot2 | 853 | 0.560 | 0.0596 | No |
| 6 | Bphl | 1027 | 0.531 | 0.0671 | No |
| 7 | Cd36 | 1179 | 0.507 | 0.0749 | No |
| 8 | Pts | 1180 | 0.506 | 0.0904 | No |
| 9 | Idh3g | 1552 | 0.463 | 0.0858 | No |
| 10 | Pcbd1 | 1685 | 0.445 | 0.0927 | No |
| 11 | Acat2 | 1709 | 0.441 | 0.1050 | No |
| 12 | Acsm3 | 1878 | 0.422 | 0.1094 | No |
| 13 | Xist | 2043 | 0.408 | 0.1136 | No |
| 14 | Gad2 | 2339 | 0.384 | 0.1105 | No |
| 15 | Eno2 | 2349 | 0.383 | 0.1217 | No |
| 16 | Acss1 | 2357 | 0.382 | 0.1330 | No |
| 17 | Gapdhs | 2499 | 0.371 | 0.1372 | No |
| 18 | Reep6 | 2640 | 0.361 | 0.1411 | No |
| 19 | Aldh1a1 | 2987 | 0.335 | 0.1339 | No |
| 20 | Rdh1 | 3103 | 0.327 | 0.1381 | No |
| 21 | Cryz | 3119 | 0.325 | 0.1473 | No |
| 22 | Hsd17b11 | 3451 | 0.303 | 0.1399 | No |
| 23 | Ncaph2 | 3505 | 0.301 | 0.1464 | No |
| 24 | Mcee | 4770 | 0.227 | 0.0897 | No |
| 25 | Mlycd | 4984 | 0.218 | 0.0857 | No |
| 26 | Hibch | 5213 | 0.207 | 0.0805 | No |
| 27 | Ltc4s | 5273 | 0.204 | 0.0837 | No |
| 28 | Gabarapl1 | 5858 | 0.173 | 0.0596 | No |
| 29 | Hadhb | 6008 | 0.165 | 0.0572 | No |
| 30 | Bckdhb | 6188 | 0.158 | 0.0530 | No |
| 31 | Cbr3 | 6291 | 0.154 | 0.0526 | No |
| 32 | Uros | 6589 | 0.142 | 0.0419 | No |
| 33 | Suclg2 | 6668 | 0.139 | 0.0423 | No |
| 34 | Sdhc | 6682 | 0.138 | 0.0458 | No |
| 35 | Serinc1 | 6706 | 0.137 | 0.0488 | No |
| 36 | Rdh11 | 6712 | 0.137 | 0.0528 | No |
| 37 | Bmpr1b | 7090 | 0.121 | 0.0375 | No |
| 38 | Gpd1 | 7172 | 0.117 | 0.0370 | No |
| 39 | Fh1 | 7195 | 0.116 | 0.0394 | No |
| 40 | Auh | 7894 | 0.088 | 0.0070 | No |
| 41 | Aldh3a2 | 7900 | 0.087 | 0.0094 | No |
| 42 | Echs1 | 8023 | 0.082 | 0.0058 | No |
| 43 | Acox1 | 8155 | 0.077 | 0.0015 | No |
| 44 | Acsl1 | 8570 | 0.062 | -0.0174 | No |
| 45 | Maoa | 8644 | 0.059 | -0.0192 | No |
| 46 | Sdha | 8708 | 0.057 | -0.0207 | No |
| 47 | Sucla2 | 8748 | 0.055 | -0.0210 | No |
| 48 | Nthl1 | 8856 | 0.051 | -0.0248 | No |
| 49 | Dlst | 9015 | 0.045 | -0.0314 | No |
| 50 | Mgll | 9210 | 0.037 | -0.0400 | No |
| 51 | Decr1 | 9573 | 0.023 | -0.0575 | No |
| 52 | Metap1 | 10534 | -0.008 | -0.1055 | No |
| 53 | Gstz1 | 10758 | -0.016 | -0.1162 | No |
| 54 | Elovl5 | 11007 | -0.025 | -0.1279 | No |
| 55 | Rap1gds1 | 11144 | -0.030 | -0.1339 | No |
| 56 | Idi1 | 11320 | -0.036 | -0.1415 | No |
| 57 | Hsd17b4 | 11371 | -0.038 | -0.1429 | No |
| 58 | Dhcr24 | 11581 | -0.045 | -0.1520 | No |
| 59 | Grhpr | 11595 | -0.045 | -0.1513 | No |
| 60 | Adipor2 | 11741 | -0.051 | -0.1571 | No |
| 61 | Ech1 | 11876 | -0.055 | -0.1621 | No |
| 62 | Blvra | 11893 | -0.056 | -0.1612 | No |
| 63 | Alad | 12317 | -0.071 | -0.1803 | No |
| 64 | Aldh9a1 | 12384 | -0.073 | -0.1814 | No |
| 65 | Etfdh | 12427 | -0.075 | -0.1812 | No |
| 66 | Pdhb | 12585 | -0.081 | -0.1866 | No |
| 67 | Nbn | 12748 | -0.087 | -0.1921 | No |
| 68 | Crat | 12920 | -0.093 | -0.1979 | No |
| 69 | Hsdl2 | 12957 | -0.094 | -0.1968 | No |
| 70 | Cpox | 13093 | -0.100 | -0.2006 | No |
| 71 | Acsl5 | 13120 | -0.101 | -0.1988 | No |
| 72 | Sms-ps | 13178 | -0.103 | -0.1985 | No |
| 73 | Ppara | 13384 | -0.110 | -0.2055 | No |
| 74 | Gcdh | 13542 | -0.115 | -0.2099 | No |
| 75 | Mdh1 | 13824 | -0.125 | -0.2202 | No |
| 76 | Prdx6 | 14147 | -0.138 | -0.2322 | No |
| 77 | Psme1 | 14163 | -0.139 | -0.2287 | No |
| 78 | Ywhah | 14230 | -0.142 | -0.2277 | No |
| 79 | Mdh2 | 14366 | -0.147 | -0.2300 | No |
| 80 | Mif | 14632 | -0.157 | -0.2385 | Yes |
| 81 | Acadm | 14651 | -0.159 | -0.2346 | Yes |
| 82 | Hmgcl | 14713 | -0.161 | -0.2328 | Yes |
| 83 | Fasn | 14800 | -0.165 | -0.2321 | Yes |
| 84 | H2az1 | 14988 | -0.173 | -0.2362 | Yes |
| 85 | Kmt5a | 15008 | -0.173 | -0.2319 | Yes |
| 86 | Slc22a5 | 15057 | -0.175 | -0.2290 | Yes |
| 87 | Hccs | 15152 | -0.179 | -0.2282 | Yes |
| 88 | Ube2l6 | 15233 | -0.182 | -0.2267 | Yes |
| 89 | Apex1 | 15439 | -0.190 | -0.2312 | Yes |
| 90 | Hmgcs1 | 15494 | -0.192 | -0.2281 | Yes |
| 91 | Eci2 | 15721 | -0.202 | -0.2333 | Yes |
| 92 | Gpd2 | 15836 | -0.207 | -0.2327 | Yes |
| 93 | D2hgdh | 15904 | -0.210 | -0.2297 | Yes |
| 94 | Odc1 | 16114 | -0.221 | -0.2335 | Yes |
| 95 | Acsl4 | 16222 | -0.226 | -0.2320 | Yes |
| 96 | Ehhadh | 16244 | -0.227 | -0.2261 | Yes |
| 97 | Aoc3 | 16256 | -0.228 | -0.2197 | Yes |
| 98 | Acads | 16417 | -0.237 | -0.2205 | Yes |
| 99 | Hmgcs2 | 16570 | -0.243 | -0.2208 | Yes |
| 100 | Pdha1 | 16718 | -0.251 | -0.2206 | Yes |
| 101 | Glul | 16907 | -0.262 | -0.2220 | Yes |
| 102 | Cpt2 | 16933 | -0.263 | -0.2153 | Yes |
| 103 | Car2 | 16954 | -0.264 | -0.2082 | Yes |
| 104 | Lgals1 | 17025 | -0.268 | -0.2036 | Yes |
| 105 | S100a10 | 17073 | -0.270 | -0.1977 | Yes |
| 106 | Acaa2 | 17084 | -0.270 | -0.1900 | Yes |
| 107 | Idh3b | 17204 | -0.277 | -0.1876 | Yes |
| 108 | Dld | 17436 | -0.292 | -0.1903 | Yes |
| 109 | Idh1 | 17515 | -0.296 | -0.1852 | Yes |
| 110 | Hsd17b10 | 17678 | -0.309 | -0.1839 | Yes |
| 111 | Nsdhl | 17680 | -0.309 | -0.1746 | Yes |
| 112 | Cbr1 | 17791 | -0.316 | -0.1705 | Yes |
| 113 | Me1 | 17827 | -0.319 | -0.1625 | Yes |
| 114 | Suclg1 | 17864 | -0.321 | -0.1546 | Yes |
| 115 | Acadl | 18011 | -0.331 | -0.1518 | Yes |
| 116 | Aldoa | 18110 | -0.338 | -0.1464 | Yes |
| 117 | Aco2 | 18132 | -0.340 | -0.1371 | Yes |
| 118 | Hsp90aa1 | 18161 | -0.342 | -0.1281 | Yes |
| 119 | Hsd17b7 | 18188 | -0.344 | -0.1189 | Yes |
| 120 | Hadh | 18242 | -0.348 | -0.1110 | Yes |
| 121 | G0s2 | 18376 | -0.361 | -0.1067 | Yes |
| 122 | Retsat | 18694 | -0.394 | -0.1106 | Yes |
| 123 | Acadvl | 18748 | -0.399 | -0.1012 | Yes |
| 124 | Erp29 | 18806 | -0.404 | -0.0917 | Yes |
| 125 | Ostc | 18841 | -0.408 | -0.0810 | Yes |
| 126 | Urod | 18856 | -0.410 | -0.0692 | Yes |
| 127 | Acot8 | 18870 | -0.411 | -0.0573 | Yes |
| 128 | Sdhd | 18902 | -0.415 | -0.0463 | Yes |
| 129 | Adsl | 18906 | -0.415 | -0.0338 | Yes |
| 130 | Hsph1 | 18973 | -0.424 | -0.0242 | Yes |
| 131 | Cpt1a | 19003 | -0.427 | -0.0127 | Yes |
| 132 | Acaa1a | 19172 | -0.446 | -0.0075 | Yes |
| 133 | Ldha | 19331 | -0.472 | -0.0011 | Yes |
| 134 | Ugdh | 19594 | -0.536 | 0.0021 | Yes |
| 135 | Eci1 | 19859 | -0.643 | 0.0084 | Yes |