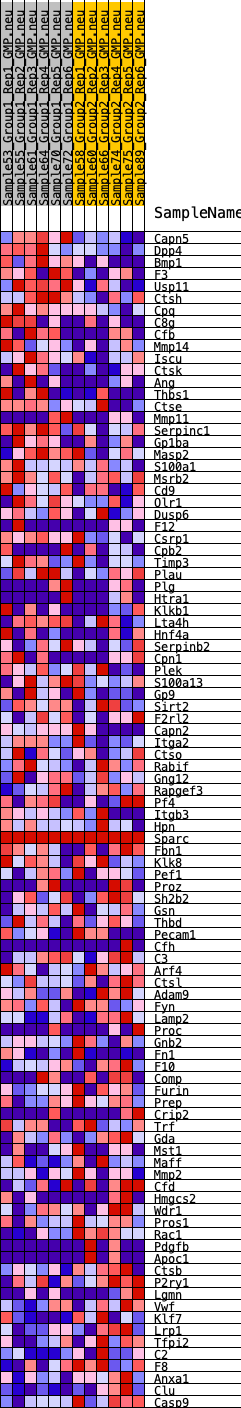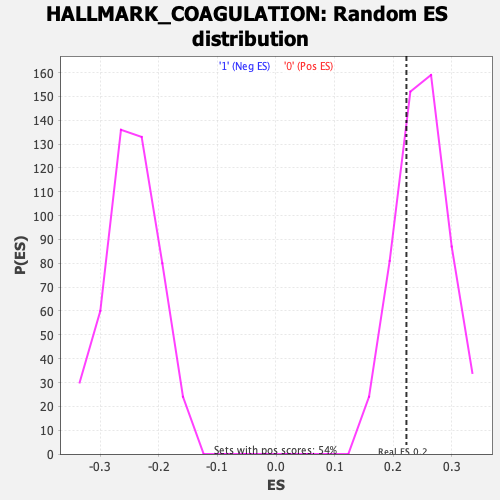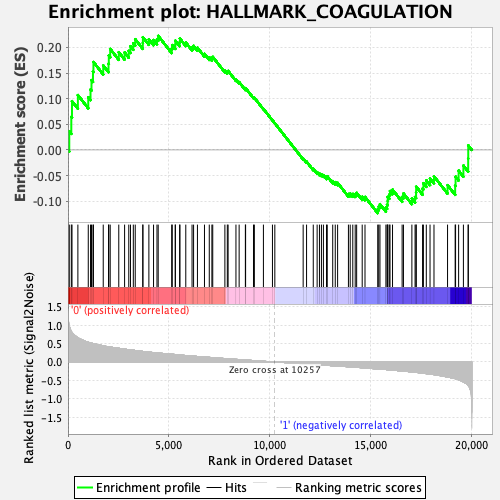
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | GMP.GMP.neu_Pheno.cls #Group1_versus_Group2.GMP.neu_Pheno.cls #Group1_versus_Group2_repos |
| Phenotype | GMP.neu_Pheno.cls#Group1_versus_Group2_repos |
| Upregulated in class | 0 |
| GeneSet | HALLMARK_COAGULATION |
| Enrichment Score (ES) | 0.22245364 |
| Normalized Enrichment Score (NES) | 0.8905515 |
| Nominal p-value | 0.7225326 |
| FDR q-value | 0.9426734 |
| FWER p-Value | 0.999 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Capn5 | 63 | 0.978 | 0.0363 | Yes |
| 2 | Dpp4 | 164 | 0.828 | 0.0647 | Yes |
| 3 | Bmp1 | 198 | 0.797 | 0.0951 | Yes |
| 4 | F3 | 490 | 0.656 | 0.1070 | Yes |
| 5 | Usp11 | 1005 | 0.533 | 0.1027 | Yes |
| 6 | Ctsh | 1117 | 0.517 | 0.1180 | Yes |
| 7 | Cpq | 1165 | 0.509 | 0.1361 | Yes |
| 8 | C8g | 1239 | 0.499 | 0.1526 | Yes |
| 9 | Cfb | 1262 | 0.496 | 0.1715 | Yes |
| 10 | Mmp14 | 1747 | 0.436 | 0.1647 | Yes |
| 11 | Iscu | 2010 | 0.411 | 0.1682 | Yes |
| 12 | Ctsk | 2024 | 0.410 | 0.1840 | Yes |
| 13 | Ang | 2093 | 0.405 | 0.1969 | Yes |
| 14 | Thbs1 | 2518 | 0.370 | 0.1906 | Yes |
| 15 | Ctse | 2807 | 0.348 | 0.1902 | Yes |
| 16 | Mmp11 | 3014 | 0.332 | 0.1932 | Yes |
| 17 | Serpinc1 | 3098 | 0.327 | 0.2023 | Yes |
| 18 | Gp1ba | 3239 | 0.318 | 0.2081 | Yes |
| 19 | Masp2 | 3334 | 0.311 | 0.2159 | Yes |
| 20 | S100a1 | 3710 | 0.285 | 0.2085 | Yes |
| 21 | Msrb2 | 3714 | 0.284 | 0.2198 | Yes |
| 22 | Cd9 | 4011 | 0.266 | 0.2157 | Yes |
| 23 | Olr1 | 4242 | 0.253 | 0.2144 | Yes |
| 24 | Dusp6 | 4413 | 0.244 | 0.2157 | Yes |
| 25 | F12 | 4473 | 0.241 | 0.2225 | Yes |
| 26 | Csrp1 | 5143 | 0.210 | 0.1974 | No |
| 27 | Cpb2 | 5170 | 0.209 | 0.2045 | No |
| 28 | Timp3 | 5321 | 0.201 | 0.2051 | No |
| 29 | Plau | 5322 | 0.201 | 0.2132 | No |
| 30 | Plg | 5539 | 0.189 | 0.2100 | No |
| 31 | Htra1 | 5549 | 0.189 | 0.2171 | No |
| 32 | Klkb1 | 5841 | 0.174 | 0.2095 | No |
| 33 | Lta4h | 6152 | 0.160 | 0.2004 | No |
| 34 | Hnf4a | 6228 | 0.157 | 0.2030 | No |
| 35 | Serpinb2 | 6419 | 0.148 | 0.1994 | No |
| 36 | Cpn1 | 6768 | 0.135 | 0.1874 | No |
| 37 | Plek | 7006 | 0.125 | 0.1805 | No |
| 38 | S100a13 | 7136 | 0.119 | 0.1788 | No |
| 39 | Gp9 | 7178 | 0.117 | 0.1815 | No |
| 40 | Sirt2 | 7780 | 0.092 | 0.1551 | No |
| 41 | F2rl2 | 7901 | 0.087 | 0.1526 | No |
| 42 | Capn2 | 7940 | 0.086 | 0.1541 | No |
| 43 | Itga2 | 8329 | 0.070 | 0.1375 | No |
| 44 | Ctso | 8487 | 0.065 | 0.1322 | No |
| 45 | Rabif | 8796 | 0.053 | 0.1189 | No |
| 46 | Gng12 | 8808 | 0.053 | 0.1205 | No |
| 47 | Rapgef3 | 9199 | 0.037 | 0.1024 | No |
| 48 | Pf4 | 9242 | 0.036 | 0.1017 | No |
| 49 | Itgb3 | 9691 | 0.020 | 0.0801 | No |
| 50 | Hpn | 10139 | 0.004 | 0.0578 | No |
| 51 | Sparc | 10257 | 0.000 | 0.0519 | No |
| 52 | Fbn1 | 11662 | -0.048 | -0.0166 | No |
| 53 | Klk8 | 11831 | -0.054 | -0.0228 | No |
| 54 | Pef1 | 12163 | -0.065 | -0.0368 | No |
| 55 | Proz | 12355 | -0.073 | -0.0435 | No |
| 56 | Sh2b2 | 12470 | -0.077 | -0.0461 | No |
| 57 | Gsn | 12568 | -0.081 | -0.0477 | No |
| 58 | Thbd | 12666 | -0.084 | -0.0492 | No |
| 59 | Pecam1 | 12821 | -0.090 | -0.0533 | No |
| 60 | Cfh | 12856 | -0.092 | -0.0513 | No |
| 61 | C3 | 13128 | -0.101 | -0.0608 | No |
| 62 | Arf4 | 13254 | -0.105 | -0.0628 | No |
| 63 | Ctsl | 13371 | -0.110 | -0.0642 | No |
| 64 | Adam9 | 13907 | -0.129 | -0.0859 | No |
| 65 | Fyn | 13995 | -0.132 | -0.0849 | No |
| 66 | Lamp2 | 14123 | -0.137 | -0.0858 | No |
| 67 | Proc | 14243 | -0.142 | -0.0860 | No |
| 68 | Gnb2 | 14315 | -0.144 | -0.0838 | No |
| 69 | Fn1 | 14590 | -0.156 | -0.0912 | No |
| 70 | F10 | 14729 | -0.162 | -0.0916 | No |
| 71 | Comp | 15360 | -0.189 | -0.1156 | No |
| 72 | Furin | 15410 | -0.189 | -0.1104 | No |
| 73 | Prep | 15471 | -0.191 | -0.1057 | No |
| 74 | Crip2 | 15766 | -0.204 | -0.1123 | No |
| 75 | Trf | 15824 | -0.206 | -0.1068 | No |
| 76 | Gda | 15853 | -0.208 | -0.0998 | No |
| 77 | Mst1 | 15856 | -0.208 | -0.0916 | No |
| 78 | Maff | 15933 | -0.211 | -0.0869 | No |
| 79 | Mmp2 | 15969 | -0.213 | -0.0800 | No |
| 80 | Cfd | 16094 | -0.219 | -0.0774 | No |
| 81 | Hmgcs2 | 16570 | -0.243 | -0.0915 | No |
| 82 | Wdr1 | 16636 | -0.246 | -0.0848 | No |
| 83 | Pros1 | 17054 | -0.269 | -0.0949 | No |
| 84 | Rac1 | 17212 | -0.278 | -0.0916 | No |
| 85 | Pdgfb | 17262 | -0.281 | -0.0827 | No |
| 86 | Apoc1 | 17267 | -0.281 | -0.0715 | No |
| 87 | Ctsb | 17584 | -0.303 | -0.0752 | No |
| 88 | P2ry1 | 17625 | -0.305 | -0.0649 | No |
| 89 | Lgmn | 17769 | -0.315 | -0.0594 | No |
| 90 | Vwf | 17954 | -0.328 | -0.0554 | No |
| 91 | Klf7 | 18150 | -0.342 | -0.0514 | No |
| 92 | Lrp1 | 18823 | -0.407 | -0.0687 | No |
| 93 | Tfpi2 | 19200 | -0.450 | -0.0694 | No |
| 94 | C2 | 19221 | -0.454 | -0.0522 | No |
| 95 | F8 | 19372 | -0.480 | -0.0403 | No |
| 96 | Anxa1 | 19609 | -0.540 | -0.0304 | No |
| 97 | Clu | 19846 | -0.634 | -0.0167 | No |
| 98 | Casp9 | 19850 | -0.637 | 0.0088 | No |