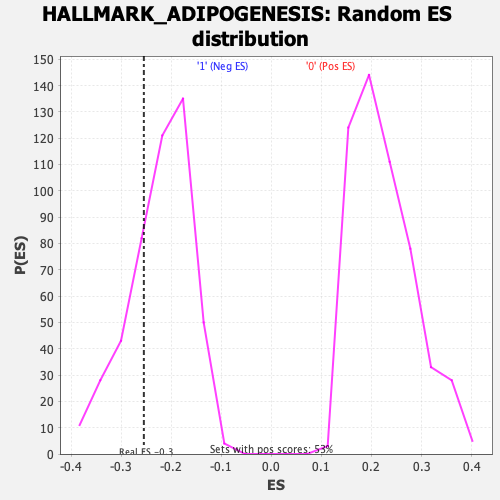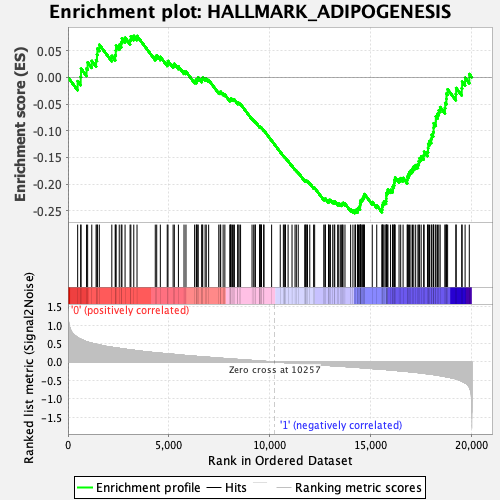
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | GMP.GMP.neu_Pheno.cls #Group1_versus_Group2.GMP.neu_Pheno.cls #Group1_versus_Group2_repos |
| Phenotype | GMP.neu_Pheno.cls#Group1_versus_Group2_repos |
| Upregulated in class | 1 |
| GeneSet | HALLMARK_ADIPOGENESIS |
| Enrichment Score (ES) | -0.25464317 |
| Normalized Enrichment Score (NES) | -1.1508633 |
| Nominal p-value | 0.23628692 |
| FDR q-value | 0.5294153 |
| FWER p-Value | 0.95 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Cyp4b1 | 479 | 0.658 | -0.0071 | No |
| 2 | Elmod3 | 630 | 0.615 | 0.0013 | No |
| 3 | Lama4 | 646 | 0.612 | 0.0163 | No |
| 4 | Tob1 | 920 | 0.549 | 0.0168 | No |
| 5 | Pparg | 974 | 0.539 | 0.0281 | No |
| 6 | Cd36 | 1179 | 0.507 | 0.0309 | No |
| 7 | Pemt | 1393 | 0.481 | 0.0326 | No |
| 8 | Angptl4 | 1437 | 0.476 | 0.0428 | No |
| 9 | Sdhb | 1457 | 0.474 | 0.0541 | No |
| 10 | Idh3g | 1552 | 0.463 | 0.0613 | No |
| 11 | Arl4a | 2173 | 0.398 | 0.0404 | No |
| 12 | Slc27a1 | 2340 | 0.384 | 0.0419 | No |
| 13 | Cavin2 | 2369 | 0.381 | 0.0504 | No |
| 14 | Uck1 | 2382 | 0.380 | 0.0596 | No |
| 15 | Angpt1 | 2541 | 0.369 | 0.0612 | No |
| 16 | Reep6 | 2640 | 0.361 | 0.0656 | No |
| 17 | Ndufa5 | 2675 | 0.358 | 0.0732 | No |
| 18 | Omd | 2830 | 0.346 | 0.0744 | No |
| 19 | Dhrs7b | 3079 | 0.329 | 0.0704 | No |
| 20 | Sult1a1 | 3117 | 0.325 | 0.0770 | No |
| 21 | Gpx3 | 3262 | 0.316 | 0.0779 | No |
| 22 | Col15a1 | 3423 | 0.305 | 0.0777 | No |
| 23 | Cavin1 | 4328 | 0.249 | 0.0386 | No |
| 24 | Itsn1 | 4399 | 0.246 | 0.0414 | No |
| 25 | Araf | 4579 | 0.235 | 0.0385 | No |
| 26 | Lipe | 4921 | 0.220 | 0.0270 | No |
| 27 | Mgst3 | 4959 | 0.219 | 0.0308 | No |
| 28 | Hibch | 5213 | 0.207 | 0.0234 | No |
| 29 | Ltc4s | 5273 | 0.204 | 0.0257 | No |
| 30 | Pgm1 | 5476 | 0.192 | 0.0205 | No |
| 31 | Lpl | 5754 | 0.178 | 0.0111 | No |
| 32 | Nkiras1 | 5850 | 0.173 | 0.0108 | No |
| 33 | Elovl6 | 6279 | 0.155 | -0.0067 | No |
| 34 | Rreb1 | 6364 | 0.151 | -0.0071 | No |
| 35 | Dnajc15 | 6368 | 0.151 | -0.0033 | No |
| 36 | Aldh2 | 6434 | 0.148 | -0.0028 | No |
| 37 | Riok3 | 6455 | 0.147 | 0.0000 | No |
| 38 | Baz2a | 6636 | 0.140 | -0.0054 | No |
| 39 | Gpam | 6645 | 0.140 | -0.0022 | No |
| 40 | Sdhc | 6682 | 0.138 | -0.0004 | No |
| 41 | Aplp2 | 6799 | 0.133 | -0.0028 | No |
| 42 | Pdcd4 | 6856 | 0.131 | -0.0023 | No |
| 43 | Slc25a1 | 6975 | 0.126 | -0.0050 | No |
| 44 | Mtarc2 | 7477 | 0.104 | -0.0275 | No |
| 45 | Samm50 | 7547 | 0.101 | -0.0284 | No |
| 46 | Gpx4 | 7572 | 0.100 | -0.0270 | No |
| 47 | Lifr | 7693 | 0.096 | -0.0306 | No |
| 48 | Bckdha | 7779 | 0.092 | -0.0325 | No |
| 49 | Echs1 | 8023 | 0.082 | -0.0426 | No |
| 50 | Dhcr7 | 8059 | 0.081 | -0.0423 | No |
| 51 | Coq3 | 8080 | 0.080 | -0.0412 | No |
| 52 | Slc5a6 | 8085 | 0.080 | -0.0393 | No |
| 53 | Acox1 | 8155 | 0.077 | -0.0408 | No |
| 54 | Adcy6 | 8218 | 0.074 | -0.0420 | No |
| 55 | Ifngr1 | 8251 | 0.072 | -0.0418 | No |
| 56 | Slc25a10 | 8419 | 0.067 | -0.0485 | No |
| 57 | Phldb1 | 8427 | 0.067 | -0.0471 | No |
| 58 | Coq9 | 8486 | 0.065 | -0.0483 | No |
| 59 | Cmbl | 8558 | 0.062 | -0.0503 | No |
| 60 | Phyh | 9127 | 0.040 | -0.0779 | No |
| 61 | Mgll | 9210 | 0.037 | -0.0811 | No |
| 62 | Dnajb9 | 9274 | 0.034 | -0.0833 | No |
| 63 | Pex14 | 9292 | 0.034 | -0.0833 | No |
| 64 | Rtn3 | 9503 | 0.026 | -0.0932 | No |
| 65 | Rnf11 | 9510 | 0.025 | -0.0929 | No |
| 66 | Bcl2l13 | 9521 | 0.025 | -0.0927 | No |
| 67 | Tst | 9559 | 0.024 | -0.0940 | No |
| 68 | Decr1 | 9573 | 0.023 | -0.0940 | No |
| 69 | Ghitm | 9596 | 0.023 | -0.0946 | No |
| 70 | Cox8a | 9704 | 0.019 | -0.0994 | No |
| 71 | Gbe1 | 9719 | 0.019 | -0.0997 | No |
| 72 | Chchd10 | 10098 | 0.006 | -0.1186 | No |
| 73 | Qdpr | 10526 | -0.008 | -0.1399 | No |
| 74 | Miga2 | 10680 | -0.013 | -0.1473 | No |
| 75 | Ppm1b | 10720 | -0.015 | -0.1489 | No |
| 76 | Sqor | 10759 | -0.016 | -0.1503 | No |
| 77 | Ndufs3 | 10785 | -0.017 | -0.1512 | No |
| 78 | Vegfb | 10915 | -0.022 | -0.1571 | No |
| 79 | Tank | 11110 | -0.028 | -0.1662 | No |
| 80 | Cyc1 | 11259 | -0.034 | -0.1727 | No |
| 81 | Atl2 | 11323 | -0.037 | -0.1750 | No |
| 82 | Cox7b | 11418 | -0.039 | -0.1787 | No |
| 83 | Adipor2 | 11741 | -0.051 | -0.1936 | No |
| 84 | Pfkl | 11771 | -0.052 | -0.1937 | No |
| 85 | Pfkfb3 | 11800 | -0.053 | -0.1937 | No |
| 86 | Slc19a1 | 11843 | -0.054 | -0.1944 | No |
| 87 | Ech1 | 11876 | -0.055 | -0.1946 | No |
| 88 | Gpat4 | 11990 | -0.059 | -0.1988 | No |
| 89 | Uqcrq | 12180 | -0.066 | -0.2066 | No |
| 90 | Por | 12232 | -0.068 | -0.2074 | No |
| 91 | Fah | 12684 | -0.085 | -0.2279 | No |
| 92 | Jagn1 | 12739 | -0.087 | -0.2284 | No |
| 93 | Enpp2 | 12766 | -0.088 | -0.2274 | No |
| 94 | Crat | 12920 | -0.093 | -0.2328 | No |
| 95 | Lpcat3 | 12921 | -0.093 | -0.2304 | No |
| 96 | Coq5 | 12960 | -0.094 | -0.2298 | No |
| 97 | Nmt1 | 13015 | -0.097 | -0.2300 | No |
| 98 | C3 | 13128 | -0.101 | -0.2331 | No |
| 99 | Etfb | 13200 | -0.104 | -0.2340 | No |
| 100 | Acly | 13229 | -0.105 | -0.2327 | No |
| 101 | Mylk | 13373 | -0.110 | -0.2370 | No |
| 102 | Rmdn3 | 13426 | -0.111 | -0.2368 | No |
| 103 | Immt | 13505 | -0.114 | -0.2378 | No |
| 104 | Dram2 | 13572 | -0.116 | -0.2381 | No |
| 105 | Nabp1 | 13608 | -0.118 | -0.2368 | No |
| 106 | Sowahc | 13640 | -0.119 | -0.2353 | No |
| 107 | Idh3a | 13736 | -0.122 | -0.2369 | No |
| 108 | Stom | 14016 | -0.133 | -0.2476 | No |
| 109 | Map4k3 | 14130 | -0.138 | -0.2497 | No |
| 110 | Ndufab1 | 14229 | -0.141 | -0.2510 | Yes |
| 111 | Mtch2 | 14261 | -0.142 | -0.2489 | Yes |
| 112 | Cat | 14359 | -0.147 | -0.2500 | Yes |
| 113 | Mdh2 | 14366 | -0.147 | -0.2465 | Yes |
| 114 | Ucp2 | 14406 | -0.148 | -0.2446 | Yes |
| 115 | Aifm1 | 14480 | -0.151 | -0.2444 | Yes |
| 116 | Ubqln1 | 14482 | -0.151 | -0.2405 | Yes |
| 117 | Scarb1 | 14489 | -0.151 | -0.2369 | Yes |
| 118 | Itih5 | 14505 | -0.152 | -0.2337 | Yes |
| 119 | Cs | 14519 | -0.153 | -0.2304 | Yes |
| 120 | Col4a1 | 14570 | -0.155 | -0.2289 | Yes |
| 121 | Reep5 | 14613 | -0.157 | -0.2270 | Yes |
| 122 | Acadm | 14651 | -0.159 | -0.2248 | Yes |
| 123 | Dhrs7 | 14680 | -0.160 | -0.2220 | Yes |
| 124 | Ptcd3 | 14701 | -0.161 | -0.2189 | Yes |
| 125 | Pim3 | 15089 | -0.177 | -0.2338 | Yes |
| 126 | Gadd45a | 15311 | -0.187 | -0.2401 | Yes |
| 127 | Rab34 | 15570 | -0.195 | -0.2481 | Yes |
| 128 | Preb | 15582 | -0.196 | -0.2435 | Yes |
| 129 | Ndufb7 | 15591 | -0.197 | -0.2389 | Yes |
| 130 | Slc1a5 | 15627 | -0.198 | -0.2355 | Yes |
| 131 | Dgat1 | 15675 | -0.200 | -0.2327 | Yes |
| 132 | Tkt | 15771 | -0.204 | -0.2322 | Yes |
| 133 | Atp1b3 | 15782 | -0.205 | -0.2274 | Yes |
| 134 | Ppp1r15b | 15786 | -0.205 | -0.2222 | Yes |
| 135 | G3bp2 | 15788 | -0.205 | -0.2170 | Yes |
| 136 | Gpd2 | 15836 | -0.207 | -0.2140 | Yes |
| 137 | Abca1 | 15867 | -0.208 | -0.2101 | Yes |
| 138 | Uqcr11 | 15996 | -0.214 | -0.2110 | Yes |
| 139 | Taldo1 | 16095 | -0.220 | -0.2103 | Yes |
| 140 | Dlat | 16106 | -0.220 | -0.2051 | Yes |
| 141 | Sorbs1 | 16157 | -0.222 | -0.2018 | Yes |
| 142 | Esrra | 16182 | -0.224 | -0.1972 | Yes |
| 143 | Stat5a | 16191 | -0.224 | -0.1918 | Yes |
| 144 | Fzd4 | 16226 | -0.226 | -0.1877 | Yes |
| 145 | Acads | 16417 | -0.237 | -0.1912 | Yes |
| 146 | Mrpl15 | 16499 | -0.240 | -0.1890 | Yes |
| 147 | Ccng2 | 16618 | -0.245 | -0.1886 | Yes |
| 148 | Ubc | 16824 | -0.256 | -0.1923 | Yes |
| 149 | Chuk | 16830 | -0.257 | -0.1859 | Yes |
| 150 | Cdkn2c | 16883 | -0.260 | -0.1818 | Yes |
| 151 | Cpt2 | 16933 | -0.263 | -0.1775 | Yes |
| 152 | Dbt | 17007 | -0.267 | -0.1742 | Yes |
| 153 | Acaa2 | 17084 | -0.270 | -0.1711 | Yes |
| 154 | Abcb8 | 17148 | -0.274 | -0.1672 | Yes |
| 155 | Uqcr10 | 17242 | -0.280 | -0.1646 | Yes |
| 156 | Cox6a1 | 17354 | -0.287 | -0.1628 | Yes |
| 157 | Mrap | 17396 | -0.290 | -0.1574 | Yes |
| 158 | Dld | 17436 | -0.292 | -0.1518 | Yes |
| 159 | Idh1 | 17515 | -0.296 | -0.1480 | Yes |
| 160 | Mccc1 | 17638 | -0.306 | -0.1463 | Yes |
| 161 | Bcl6 | 17660 | -0.308 | -0.1393 | Yes |
| 162 | Me1 | 17827 | -0.319 | -0.1395 | Yes |
| 163 | Apoe | 17856 | -0.321 | -0.1326 | Yes |
| 164 | Suclg1 | 17864 | -0.321 | -0.1246 | Yes |
| 165 | Ephx2 | 17927 | -0.326 | -0.1193 | Yes |
| 166 | Acadl | 18011 | -0.331 | -0.1149 | Yes |
| 167 | Cmpk1 | 18038 | -0.334 | -0.1076 | Yes |
| 168 | Aldoa | 18110 | -0.338 | -0.1024 | Yes |
| 169 | Aco2 | 18132 | -0.340 | -0.0947 | Yes |
| 170 | Cd151 | 18136 | -0.341 | -0.0860 | Yes |
| 171 | Gphn | 18241 | -0.348 | -0.0823 | Yes |
| 172 | Hadh | 18242 | -0.348 | -0.0732 | Yes |
| 173 | Ywhag | 18327 | -0.356 | -0.0683 | Yes |
| 174 | Uqcrc1 | 18388 | -0.362 | -0.0619 | Yes |
| 175 | Agpat3 | 18456 | -0.370 | -0.0557 | Yes |
| 176 | Retsat | 18694 | -0.394 | -0.0575 | Yes |
| 177 | Cd302 | 18707 | -0.395 | -0.0479 | Yes |
| 178 | Esyt1 | 18765 | -0.400 | -0.0404 | Yes |
| 179 | Scp2 | 18769 | -0.401 | -0.0301 | Yes |
| 180 | Ak2 | 18826 | -0.407 | -0.0224 | Yes |
| 181 | Sod1 | 19236 | -0.456 | -0.0312 | Yes |
| 182 | Grpel1 | 19249 | -0.460 | -0.0199 | Yes |
| 183 | Ptger3 | 19527 | -0.519 | -0.0205 | Yes |
| 184 | Plin2 | 19545 | -0.522 | -0.0078 | Yes |
| 185 | Prdx3 | 19693 | -0.563 | -0.0007 | Yes |
| 186 | Ddt | 19901 | -0.673 | 0.0063 | Yes |