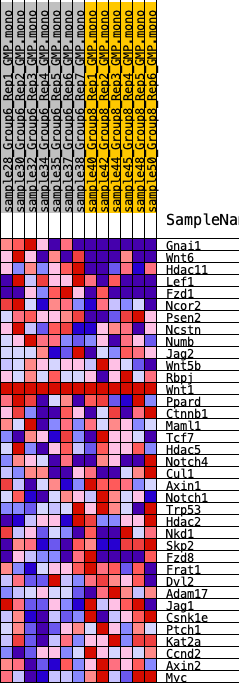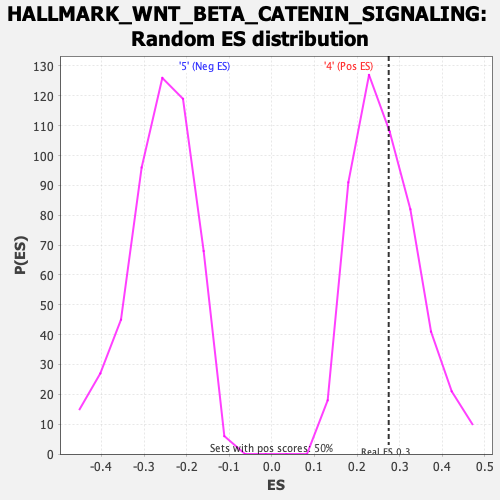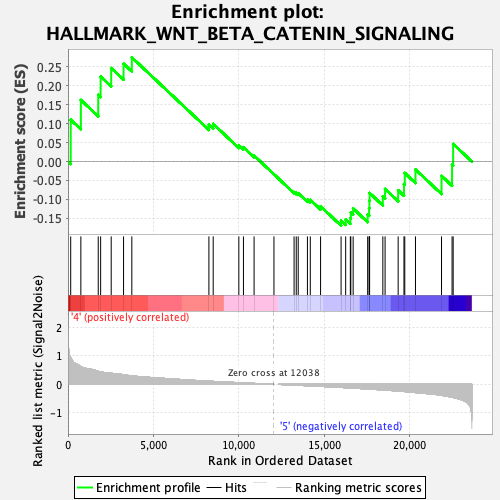
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | GMP.GMP.mono_Pheno.cls #Group6_versus_Group8.GMP.mono_Pheno.cls #Group6_versus_Group8_repos |
| Phenotype | GMP.mono_Pheno.cls#Group6_versus_Group8_repos |
| Upregulated in class | 4 |
| GeneSet | HALLMARK_WNT_BETA_CATENIN_SIGNALING |
| Enrichment Score (ES) | 0.27421856 |
| Normalized Enrichment Score (NES) | 1.0236528 |
| Nominal p-value | 0.4176707 |
| FDR q-value | 0.56399786 |
| FWER p-Value | 0.997 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Gnai1 | 158 | 0.940 | 0.1101 | Yes |
| 2 | Wnt6 | 752 | 0.621 | 0.1621 | Yes |
| 3 | Hdac11 | 1769 | 0.456 | 0.1756 | Yes |
| 4 | Lef1 | 1909 | 0.437 | 0.2240 | Yes |
| 5 | Fzd1 | 2527 | 0.389 | 0.2462 | Yes |
| 6 | Ncor2 | 3250 | 0.339 | 0.2577 | Yes |
| 7 | Psen2 | 3734 | 0.298 | 0.2742 | Yes |
| 8 | Ncstn | 8243 | 0.111 | 0.0969 | No |
| 9 | Numb | 8494 | 0.103 | 0.0990 | No |
| 10 | Jag2 | 9994 | 0.059 | 0.0428 | No |
| 11 | Wnt5b | 10264 | 0.051 | 0.0377 | No |
| 12 | Rbpj | 10886 | 0.034 | 0.0156 | No |
| 13 | Wnt1 | 12049 | 0.000 | -0.0337 | No |
| 14 | Ppard | 13226 | -0.024 | -0.0805 | No |
| 15 | Ctnnb1 | 13353 | -0.028 | -0.0824 | No |
| 16 | Maml1 | 13468 | -0.031 | -0.0834 | No |
| 17 | Tcf7 | 14007 | -0.047 | -0.1004 | No |
| 18 | Hdac5 | 14175 | -0.052 | -0.1011 | No |
| 19 | Notch4 | 14776 | -0.069 | -0.1180 | No |
| 20 | Cul1 | 15976 | -0.106 | -0.1557 | No |
| 21 | Axin1 | 16244 | -0.115 | -0.1527 | No |
| 22 | Notch1 | 16517 | -0.124 | -0.1488 | No |
| 23 | Trp53 | 16545 | -0.126 | -0.1343 | No |
| 24 | Hdac2 | 16682 | -0.131 | -0.1238 | No |
| 25 | Nkd1 | 17531 | -0.161 | -0.1398 | No |
| 26 | Skp2 | 17615 | -0.164 | -0.1228 | No |
| 27 | Fzd8 | 17634 | -0.165 | -0.1031 | No |
| 28 | Frat1 | 17645 | -0.166 | -0.0829 | No |
| 29 | Dvl2 | 18419 | -0.194 | -0.0916 | No |
| 30 | Adam17 | 18550 | -0.200 | -0.0723 | No |
| 31 | Jag1 | 19315 | -0.232 | -0.0759 | No |
| 32 | Csnk1e | 19655 | -0.248 | -0.0594 | No |
| 33 | Ptch1 | 19702 | -0.251 | -0.0302 | No |
| 34 | Kat2a | 20328 | -0.287 | -0.0211 | No |
| 35 | Ccnd2 | 21852 | -0.384 | -0.0379 | No |
| 36 | Axin2 | 22469 | -0.450 | -0.0081 | No |
| 37 | Myc | 22533 | -0.459 | 0.0463 | No |