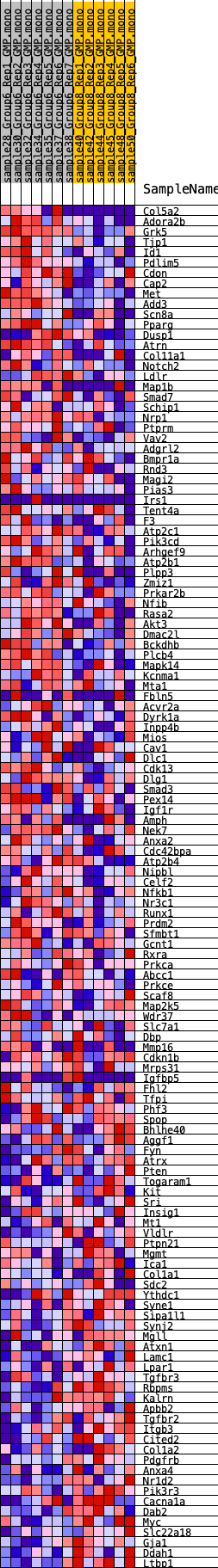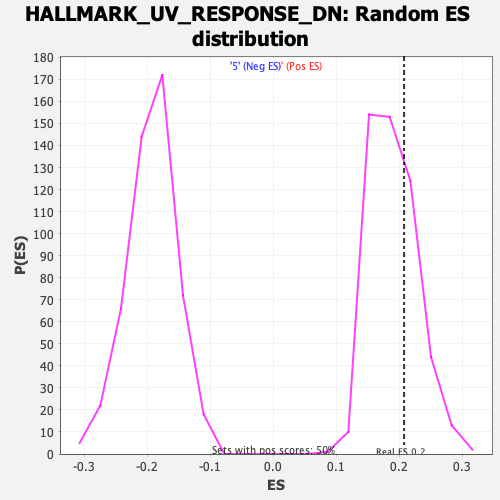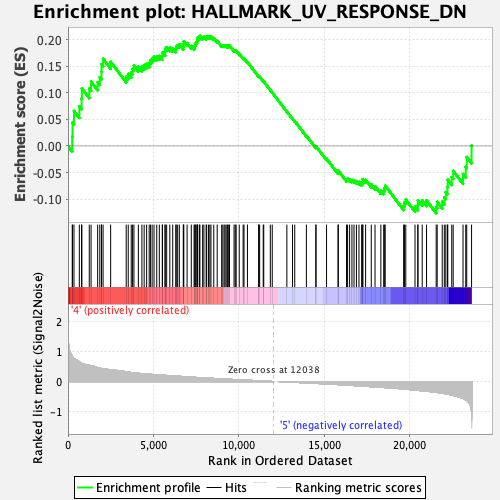
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | GMP.GMP.mono_Pheno.cls #Group6_versus_Group8.GMP.mono_Pheno.cls #Group6_versus_Group8_repos |
| Phenotype | GMP.mono_Pheno.cls#Group6_versus_Group8_repos |
| Upregulated in class | 4 |
| GeneSet | HALLMARK_UV_RESPONSE_DN |
| Enrichment Score (ES) | 0.20768337 |
| Normalized Enrichment Score (NES) | 1.0907702 |
| Nominal p-value | 0.31936127 |
| FDR q-value | 0.7919115 |
| FWER p-Value | 0.986 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Col5a2 | 247 | 0.842 | 0.0175 | Yes |
| 2 | Adora2b | 264 | 0.828 | 0.0444 | Yes |
| 3 | Grk5 | 350 | 0.763 | 0.0661 | Yes |
| 4 | Tjp1 | 662 | 0.657 | 0.0747 | Yes |
| 5 | Id1 | 793 | 0.606 | 0.0894 | Yes |
| 6 | Pdlim5 | 811 | 0.601 | 0.1086 | Yes |
| 7 | Cdon | 1244 | 0.540 | 0.1082 | Yes |
| 8 | Cap2 | 1350 | 0.527 | 0.1213 | Yes |
| 9 | Met | 1735 | 0.462 | 0.1203 | Yes |
| 10 | Add3 | 1860 | 0.443 | 0.1298 | Yes |
| 11 | Scn8a | 1958 | 0.432 | 0.1400 | Yes |
| 12 | Pparg | 1962 | 0.431 | 0.1542 | Yes |
| 13 | Dusp1 | 2053 | 0.421 | 0.1644 | Yes |
| 14 | Atrn | 2495 | 0.394 | 0.1587 | Yes |
| 15 | Col11a1 | 3405 | 0.322 | 0.1308 | Yes |
| 16 | Notch2 | 3524 | 0.312 | 0.1361 | Yes |
| 17 | Ldlr | 3699 | 0.300 | 0.1387 | Yes |
| 18 | Map1b | 3778 | 0.296 | 0.1452 | Yes |
| 19 | Smad7 | 3857 | 0.290 | 0.1515 | Yes |
| 20 | Schip1 | 4116 | 0.274 | 0.1497 | Yes |
| 21 | Nrp1 | 4324 | 0.264 | 0.1496 | Yes |
| 22 | Ptprm | 4455 | 0.260 | 0.1527 | Yes |
| 23 | Vav2 | 4600 | 0.250 | 0.1549 | Yes |
| 24 | Adgrl2 | 4749 | 0.246 | 0.1568 | Yes |
| 25 | Bmpr1a | 4821 | 0.241 | 0.1618 | Yes |
| 26 | Rnd3 | 4921 | 0.235 | 0.1654 | Yes |
| 27 | Magi2 | 5032 | 0.229 | 0.1683 | Yes |
| 28 | Pias3 | 5191 | 0.223 | 0.1690 | Yes |
| 29 | Irs1 | 5342 | 0.220 | 0.1700 | Yes |
| 30 | Tent4a | 5516 | 0.214 | 0.1697 | Yes |
| 31 | F3 | 5527 | 0.213 | 0.1764 | Yes |
| 32 | Atp2c1 | 5677 | 0.206 | 0.1769 | Yes |
| 33 | Pik3cd | 5684 | 0.205 | 0.1835 | Yes |
| 34 | Arhgef9 | 5771 | 0.201 | 0.1865 | Yes |
| 35 | Atp2b1 | 5956 | 0.197 | 0.1852 | Yes |
| 36 | Plpp3 | 6126 | 0.190 | 0.1843 | Yes |
| 37 | Zmiz1 | 6307 | 0.181 | 0.1827 | Yes |
| 38 | Prkar2b | 6329 | 0.181 | 0.1878 | Yes |
| 39 | Nfib | 6415 | 0.177 | 0.1901 | Yes |
| 40 | Rasa2 | 6513 | 0.174 | 0.1917 | Yes |
| 41 | Akt3 | 6743 | 0.165 | 0.1875 | Yes |
| 42 | Dmac2l | 6763 | 0.164 | 0.1921 | Yes |
| 43 | Bckdhb | 6771 | 0.164 | 0.1973 | Yes |
| 44 | Plcb4 | 6967 | 0.156 | 0.1942 | Yes |
| 45 | Mapk14 | 7212 | 0.146 | 0.1886 | Yes |
| 46 | Kcnma1 | 7376 | 0.140 | 0.1864 | Yes |
| 47 | Mta1 | 7418 | 0.139 | 0.1892 | Yes |
| 48 | Fbln5 | 7442 | 0.138 | 0.1928 | Yes |
| 49 | Acvr2a | 7489 | 0.136 | 0.1954 | Yes |
| 50 | Dyrk1a | 7546 | 0.134 | 0.1974 | Yes |
| 51 | Inpp4b | 7567 | 0.133 | 0.2010 | Yes |
| 52 | Mios | 7586 | 0.132 | 0.2047 | Yes |
| 53 | Cav1 | 7686 | 0.129 | 0.2047 | Yes |
| 54 | Dlc1 | 7717 | 0.127 | 0.2077 | Yes |
| 55 | Cdk13 | 7876 | 0.122 | 0.2050 | No |
| 56 | Dlg1 | 7950 | 0.120 | 0.2059 | No |
| 57 | Smad3 | 8080 | 0.116 | 0.2043 | No |
| 58 | Pex14 | 8112 | 0.114 | 0.2068 | No |
| 59 | Igf1r | 8217 | 0.112 | 0.2061 | No |
| 60 | Amph | 8298 | 0.109 | 0.2063 | No |
| 61 | Nek7 | 8364 | 0.107 | 0.2071 | No |
| 62 | Anxa2 | 8528 | 0.102 | 0.2035 | No |
| 63 | Cdc42bpa | 8728 | 0.095 | 0.1982 | No |
| 64 | Atp2b4 | 8990 | 0.088 | 0.1900 | No |
| 65 | Nipbl | 9060 | 0.086 | 0.1900 | No |
| 66 | Celf2 | 9136 | 0.084 | 0.1896 | No |
| 67 | Nfkb1 | 9196 | 0.082 | 0.1898 | No |
| 68 | Nr3c1 | 9293 | 0.080 | 0.1883 | No |
| 69 | Runx1 | 9323 | 0.078 | 0.1897 | No |
| 70 | Prdm2 | 9401 | 0.076 | 0.1890 | No |
| 71 | Sfmbt1 | 9448 | 0.074 | 0.1895 | No |
| 72 | Gcnt1 | 9715 | 0.067 | 0.1804 | No |
| 73 | Rxra | 9785 | 0.065 | 0.1796 | No |
| 74 | Prkca | 9838 | 0.063 | 0.1795 | No |
| 75 | Abcc1 | 10017 | 0.058 | 0.1738 | No |
| 76 | Prkce | 10246 | 0.051 | 0.1658 | No |
| 77 | Scaf8 | 10297 | 0.050 | 0.1654 | No |
| 78 | Map2k5 | 10498 | 0.044 | 0.1583 | No |
| 79 | Wdr37 | 11159 | 0.025 | 0.1311 | No |
| 80 | Slc7a1 | 11160 | 0.025 | 0.1319 | No |
| 81 | Dbp | 11212 | 0.024 | 0.1305 | No |
| 82 | Mmp16 | 11420 | 0.018 | 0.1223 | No |
| 83 | Cdkn1b | 11452 | 0.017 | 0.1215 | No |
| 84 | Mrps31 | 11836 | 0.006 | 0.1054 | No |
| 85 | Igfbp5 | 11949 | 0.002 | 0.1007 | No |
| 86 | Fhl2 | 12804 | -0.013 | 0.0648 | No |
| 87 | Tfpi | 13135 | -0.022 | 0.0515 | No |
| 88 | Phf3 | 13265 | -0.026 | 0.0468 | No |
| 89 | Spop | 13946 | -0.045 | 0.0194 | No |
| 90 | Bhlhe40 | 14500 | -0.060 | -0.0021 | No |
| 91 | Aggf1 | 14509 | -0.060 | -0.0005 | No |
| 92 | Fyn | 15125 | -0.080 | -0.0240 | No |
| 93 | Atrx | 15796 | -0.100 | -0.0492 | No |
| 94 | Pten | 15814 | -0.101 | -0.0466 | No |
| 95 | Togaram1 | 16294 | -0.117 | -0.0631 | No |
| 96 | Kit | 16357 | -0.119 | -0.0618 | No |
| 97 | Sri | 16475 | -0.123 | -0.0627 | No |
| 98 | Insig1 | 16611 | -0.129 | -0.0641 | No |
| 99 | Mt1 | 16733 | -0.133 | -0.0649 | No |
| 100 | Vldlr | 16873 | -0.137 | -0.0662 | No |
| 101 | Ptpn21 | 17022 | -0.142 | -0.0678 | No |
| 102 | Mgmt | 17178 | -0.149 | -0.0694 | No |
| 103 | Ica1 | 17240 | -0.151 | -0.0670 | No |
| 104 | Col1a1 | 17248 | -0.151 | -0.0623 | No |
| 105 | Sdc2 | 17411 | -0.157 | -0.0640 | No |
| 106 | Ythdc1 | 17741 | -0.169 | -0.0723 | No |
| 107 | Syne1 | 17962 | -0.178 | -0.0758 | No |
| 108 | Sipa1l1 | 18303 | -0.189 | -0.0840 | No |
| 109 | Synj2 | 18471 | -0.196 | -0.0846 | No |
| 110 | Mgll | 18506 | -0.198 | -0.0794 | No |
| 111 | Atxn1 | 18557 | -0.200 | -0.0749 | No |
| 112 | Lamc1 | 19636 | -0.247 | -0.1126 | No |
| 113 | Lpar1 | 19683 | -0.250 | -0.1062 | No |
| 114 | Tgfbr3 | 19762 | -0.254 | -0.1011 | No |
| 115 | Rbpms | 20301 | -0.285 | -0.1145 | No |
| 116 | Kalrn | 20465 | -0.292 | -0.1117 | No |
| 117 | Apbb2 | 20473 | -0.293 | -0.1023 | No |
| 118 | Tgfbr2 | 20722 | -0.307 | -0.1026 | No |
| 119 | Itgb3 | 20975 | -0.321 | -0.1027 | No |
| 120 | Cited2 | 21539 | -0.357 | -0.1148 | No |
| 121 | Col1a2 | 21598 | -0.361 | -0.1052 | No |
| 122 | Pdgfrb | 21894 | -0.388 | -0.1049 | No |
| 123 | Anxa4 | 22016 | -0.399 | -0.0968 | No |
| 124 | Nr1d2 | 22103 | -0.408 | -0.0869 | No |
| 125 | Pik3r3 | 22198 | -0.419 | -0.0769 | No |
| 126 | Cacna1a | 22219 | -0.421 | -0.0638 | No |
| 127 | Dab2 | 22451 | -0.448 | -0.0587 | No |
| 128 | Myc | 22533 | -0.459 | -0.0469 | No |
| 129 | Slc22a18 | 23110 | -0.559 | -0.0528 | No |
| 130 | Gja1 | 23269 | -0.609 | -0.0393 | No |
| 131 | Ddah1 | 23327 | -0.627 | -0.0208 | No |
| 132 | Ltbp1 | 23610 | -1.005 | 0.0006 | No |