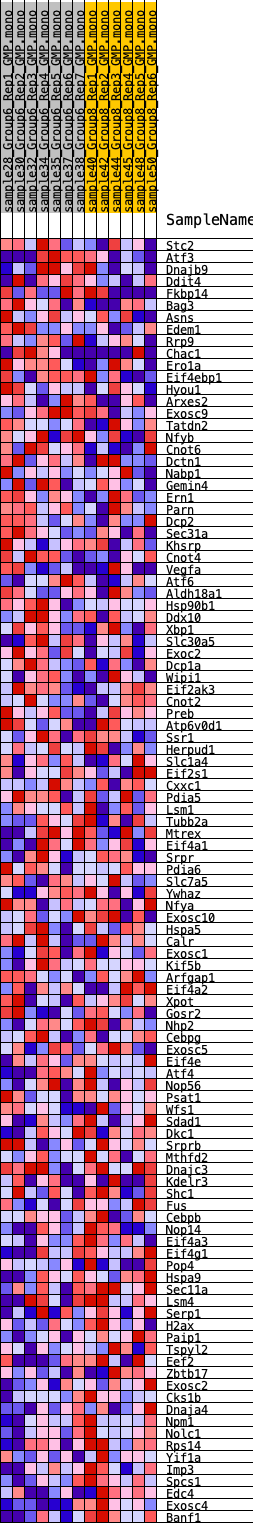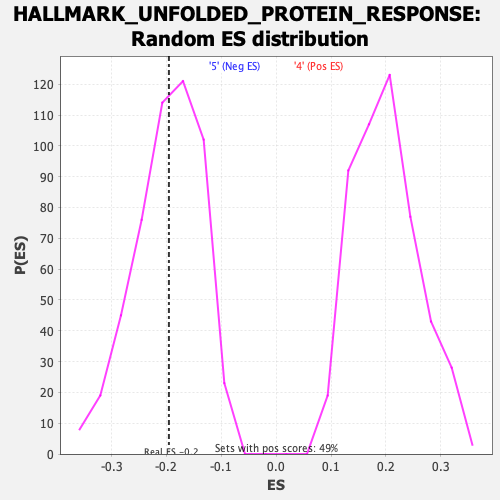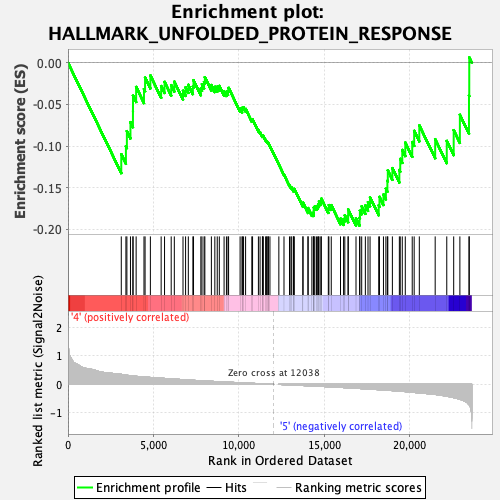
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | GMP.GMP.mono_Pheno.cls #Group6_versus_Group8.GMP.mono_Pheno.cls #Group6_versus_Group8_repos |
| Phenotype | GMP.mono_Pheno.cls#Group6_versus_Group8_repos |
| Upregulated in class | 5 |
| GeneSet | HALLMARK_UNFOLDED_PROTEIN_RESPONSE |
| Enrichment Score (ES) | -0.19542317 |
| Normalized Enrichment Score (NES) | -0.9877542 |
| Nominal p-value | 0.47047246 |
| FDR q-value | 0.8093713 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Stc2 | 3117 | 0.351 | -0.1099 | No |
| 2 | Atf3 | 3388 | 0.324 | -0.1004 | No |
| 3 | Dnajb9 | 3445 | 0.319 | -0.0821 | No |
| 4 | Ddit4 | 3654 | 0.303 | -0.0713 | No |
| 5 | Fkbp14 | 3800 | 0.294 | -0.0584 | No |
| 6 | Bag3 | 3802 | 0.294 | -0.0395 | No |
| 7 | Asns | 3986 | 0.281 | -0.0290 | No |
| 8 | Edem1 | 4445 | 0.260 | -0.0317 | No |
| 9 | Rrp9 | 4509 | 0.256 | -0.0178 | No |
| 10 | Chac1 | 4819 | 0.241 | -0.0153 | No |
| 11 | Ero1a | 5449 | 0.218 | -0.0280 | No |
| 12 | Eif4ebp1 | 5648 | 0.207 | -0.0230 | No |
| 13 | Hyou1 | 6037 | 0.194 | -0.0269 | No |
| 14 | Arxes2 | 6220 | 0.185 | -0.0227 | No |
| 15 | Exosc9 | 6728 | 0.165 | -0.0335 | No |
| 16 | Tatdn2 | 6879 | 0.160 | -0.0296 | No |
| 17 | Nfyb | 7039 | 0.153 | -0.0265 | No |
| 18 | Cnot6 | 7306 | 0.142 | -0.0286 | No |
| 19 | Dctn1 | 7343 | 0.141 | -0.0210 | No |
| 20 | Nabp1 | 7772 | 0.126 | -0.0310 | No |
| 21 | Gemin4 | 7838 | 0.124 | -0.0258 | No |
| 22 | Ern1 | 7960 | 0.120 | -0.0232 | No |
| 23 | Parn | 8006 | 0.118 | -0.0174 | No |
| 24 | Dcp2 | 8388 | 0.107 | -0.0268 | No |
| 25 | Sec31a | 8590 | 0.100 | -0.0289 | No |
| 26 | Khsrp | 8731 | 0.095 | -0.0286 | No |
| 27 | Cnot4 | 8852 | 0.092 | -0.0278 | No |
| 28 | Vegfa | 9131 | 0.084 | -0.0342 | No |
| 29 | Atf6 | 9269 | 0.080 | -0.0348 | No |
| 30 | Aldh18a1 | 9360 | 0.077 | -0.0337 | No |
| 31 | Hsp90b1 | 9395 | 0.076 | -0.0302 | No |
| 32 | Ddx10 | 10068 | 0.057 | -0.0551 | No |
| 33 | Xbp1 | 10182 | 0.053 | -0.0564 | No |
| 34 | Slc30a5 | 10202 | 0.053 | -0.0538 | No |
| 35 | Exoc2 | 10270 | 0.051 | -0.0534 | No |
| 36 | Dcp1a | 10390 | 0.047 | -0.0554 | No |
| 37 | Wipi1 | 10755 | 0.037 | -0.0685 | No |
| 38 | Eif2ak3 | 10797 | 0.036 | -0.0679 | No |
| 39 | Cnot2 | 11139 | 0.026 | -0.0807 | No |
| 40 | Preb | 11236 | 0.023 | -0.0833 | No |
| 41 | Atp6v0d1 | 11374 | 0.019 | -0.0879 | No |
| 42 | Ssr1 | 11385 | 0.019 | -0.0872 | No |
| 43 | Herpud1 | 11427 | 0.017 | -0.0878 | No |
| 44 | Slc1a4 | 11561 | 0.013 | -0.0926 | No |
| 45 | Eif2s1 | 11619 | 0.012 | -0.0943 | No |
| 46 | Cxxc1 | 11648 | 0.011 | -0.0947 | No |
| 47 | Pdia5 | 11710 | 0.009 | -0.0967 | No |
| 48 | Lsm1 | 11723 | 0.009 | -0.0967 | No |
| 49 | Tubb2a | 11814 | 0.006 | -0.1001 | No |
| 50 | Mtrex | 12335 | -0.001 | -0.1222 | No |
| 51 | Eif4a1 | 12636 | -0.008 | -0.1344 | No |
| 52 | Srpr | 12958 | -0.017 | -0.1469 | No |
| 53 | Pdia6 | 13041 | -0.019 | -0.1492 | No |
| 54 | Slc7a5 | 13090 | -0.020 | -0.1499 | No |
| 55 | Ywhaz | 13195 | -0.023 | -0.1528 | No |
| 56 | Nfya | 13198 | -0.024 | -0.1514 | No |
| 57 | Exosc10 | 13239 | -0.025 | -0.1515 | No |
| 58 | Hspa5 | 13744 | -0.039 | -0.1704 | No |
| 59 | Calr | 13746 | -0.039 | -0.1679 | No |
| 60 | Exosc1 | 14040 | -0.048 | -0.1772 | No |
| 61 | Kif5b | 14046 | -0.048 | -0.1744 | No |
| 62 | Arfgap1 | 14267 | -0.054 | -0.1802 | No |
| 63 | Eif4a2 | 14357 | -0.057 | -0.1803 | No |
| 64 | Xpot | 14378 | -0.057 | -0.1775 | No |
| 65 | Gosr2 | 14379 | -0.057 | -0.1738 | No |
| 66 | Nhp2 | 14438 | -0.059 | -0.1725 | No |
| 67 | Cebpg | 14535 | -0.061 | -0.1726 | No |
| 68 | Exosc5 | 14606 | -0.063 | -0.1715 | No |
| 69 | Eif4e | 14656 | -0.065 | -0.1694 | No |
| 70 | Atf4 | 14686 | -0.066 | -0.1663 | No |
| 71 | Nop56 | 14792 | -0.069 | -0.1663 | No |
| 72 | Psat1 | 14822 | -0.070 | -0.1630 | No |
| 73 | Wfs1 | 15229 | -0.083 | -0.1749 | No |
| 74 | Sdad1 | 15263 | -0.085 | -0.1708 | No |
| 75 | Dkc1 | 15401 | -0.089 | -0.1709 | No |
| 76 | Srprb | 15942 | -0.105 | -0.1871 | No |
| 77 | Mthfd2 | 16122 | -0.111 | -0.1875 | Yes |
| 78 | Dnajc3 | 16194 | -0.113 | -0.1832 | Yes |
| 79 | Kdelr3 | 16389 | -0.120 | -0.1837 | Yes |
| 80 | Shc1 | 16391 | -0.120 | -0.1760 | Yes |
| 81 | Fus | 16849 | -0.136 | -0.1866 | Yes |
| 82 | Cebpb | 17058 | -0.144 | -0.1861 | Yes |
| 83 | Nop14 | 17077 | -0.144 | -0.1775 | Yes |
| 84 | Eif4a3 | 17182 | -0.149 | -0.1723 | Yes |
| 85 | Eif4g1 | 17396 | -0.157 | -0.1712 | Yes |
| 86 | Pop4 | 17551 | -0.162 | -0.1673 | Yes |
| 87 | Hspa9 | 17671 | -0.167 | -0.1616 | Yes |
| 88 | Sec11a | 18178 | -0.185 | -0.1712 | Yes |
| 89 | Lsm4 | 18222 | -0.186 | -0.1609 | Yes |
| 90 | Serp1 | 18455 | -0.196 | -0.1581 | Yes |
| 91 | H2ax | 18597 | -0.201 | -0.1511 | Yes |
| 92 | Paip1 | 18685 | -0.205 | -0.1415 | Yes |
| 93 | Tspyl2 | 18710 | -0.206 | -0.1292 | Yes |
| 94 | Eef2 | 18977 | -0.218 | -0.1265 | Yes |
| 95 | Zbtb17 | 19385 | -0.235 | -0.1285 | Yes |
| 96 | Exosc2 | 19437 | -0.238 | -0.1153 | Yes |
| 97 | Cks1b | 19556 | -0.243 | -0.1047 | Yes |
| 98 | Dnaja4 | 19732 | -0.252 | -0.0958 | Yes |
| 99 | Npm1 | 20138 | -0.276 | -0.0951 | Yes |
| 100 | Nolc1 | 20252 | -0.282 | -0.0817 | Yes |
| 101 | Rps14 | 20553 | -0.297 | -0.0752 | Yes |
| 102 | Yif1a | 21481 | -0.353 | -0.0918 | Yes |
| 103 | Imp3 | 22159 | -0.414 | -0.0938 | Yes |
| 104 | Spcs1 | 22564 | -0.464 | -0.0809 | Yes |
| 105 | Edc4 | 22926 | -0.523 | -0.0624 | Yes |
| 106 | Exosc4 | 23459 | -0.705 | -0.0395 | Yes |
| 107 | Banf1 | 23475 | -0.718 | 0.0063 | Yes |